#
#
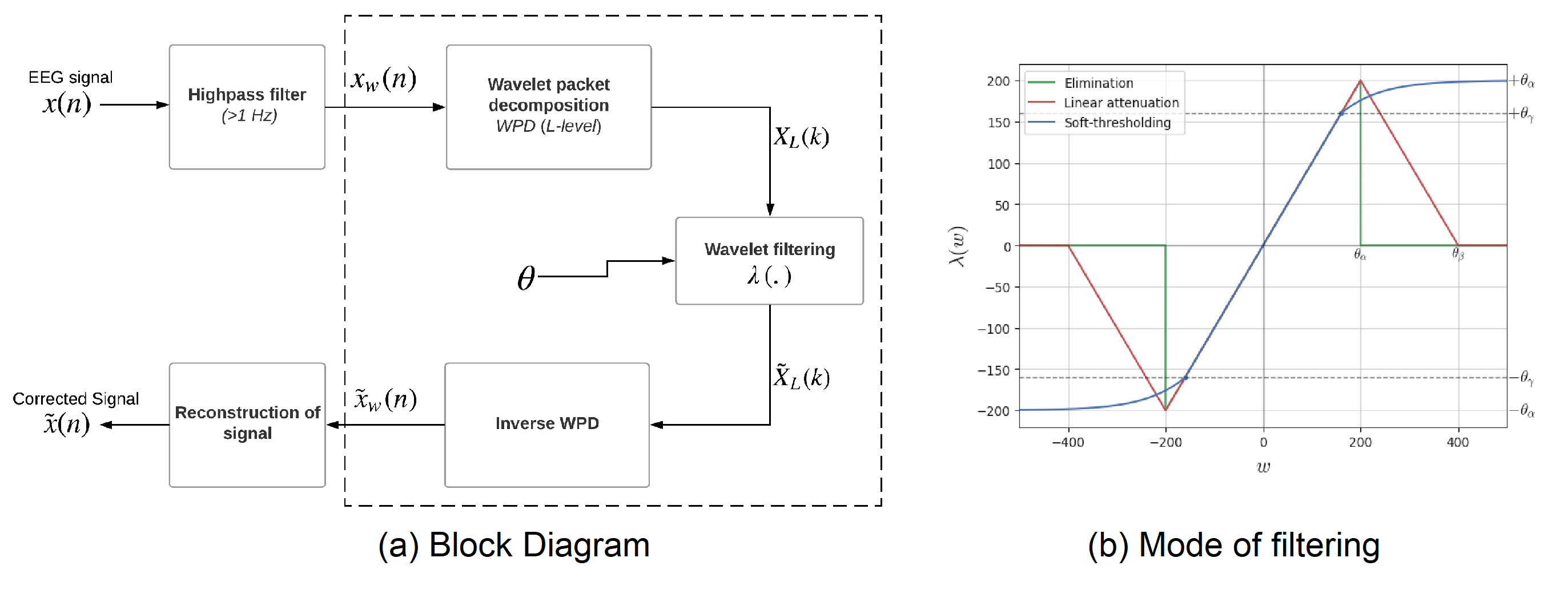
Fig 1: ATAR Algorithm Block Diagram and Mode of filtering
#
 #
#  #
# 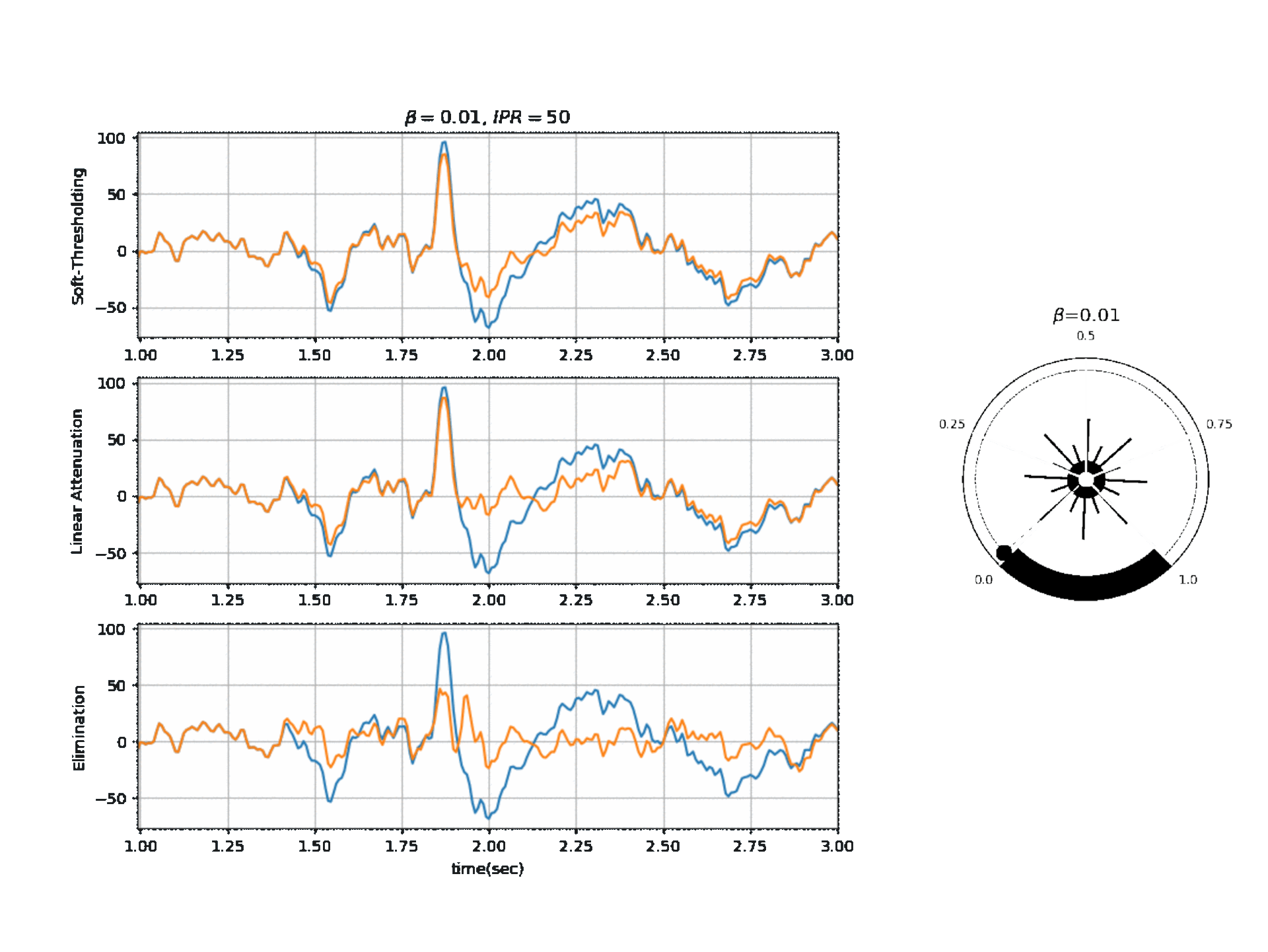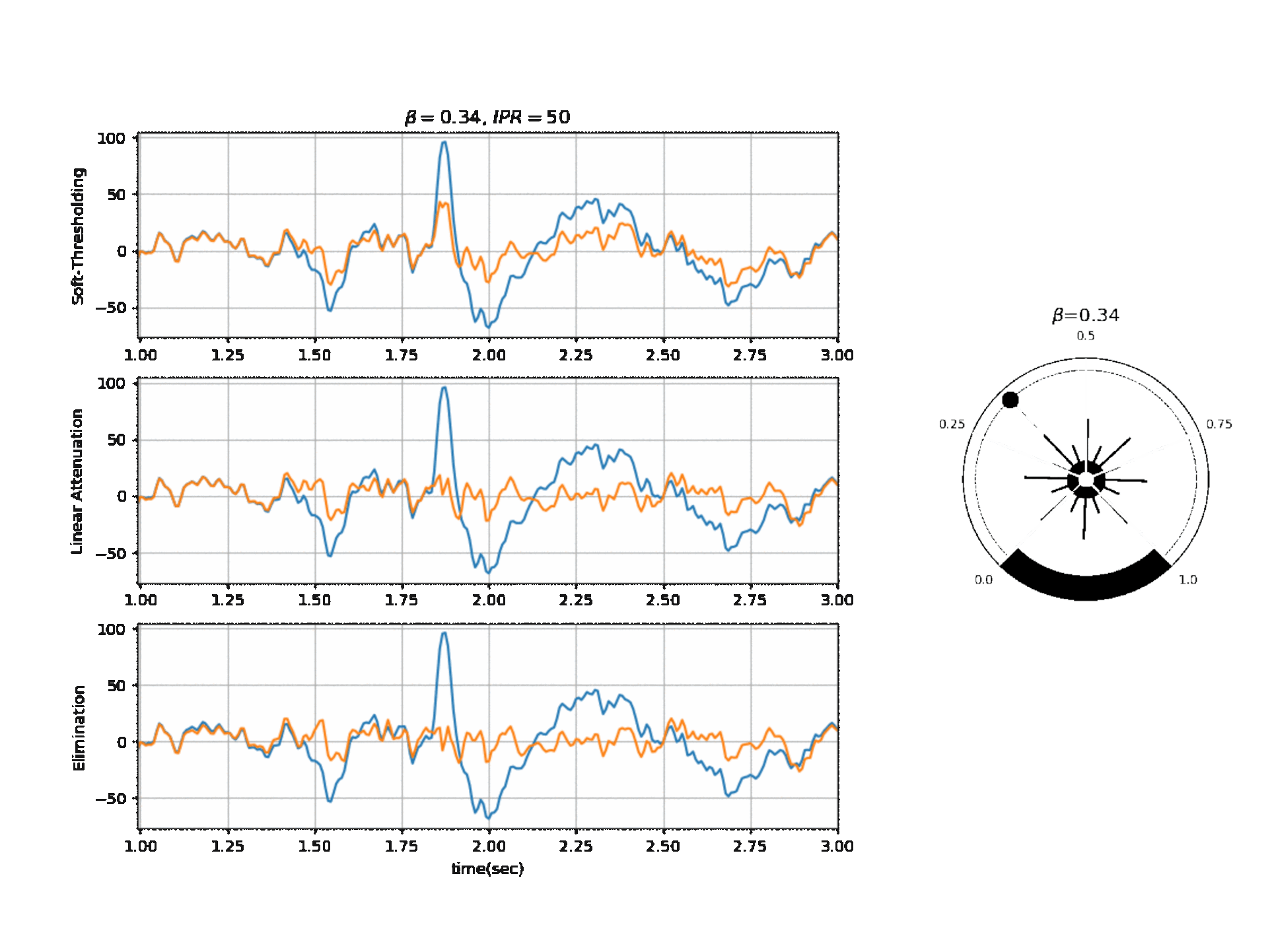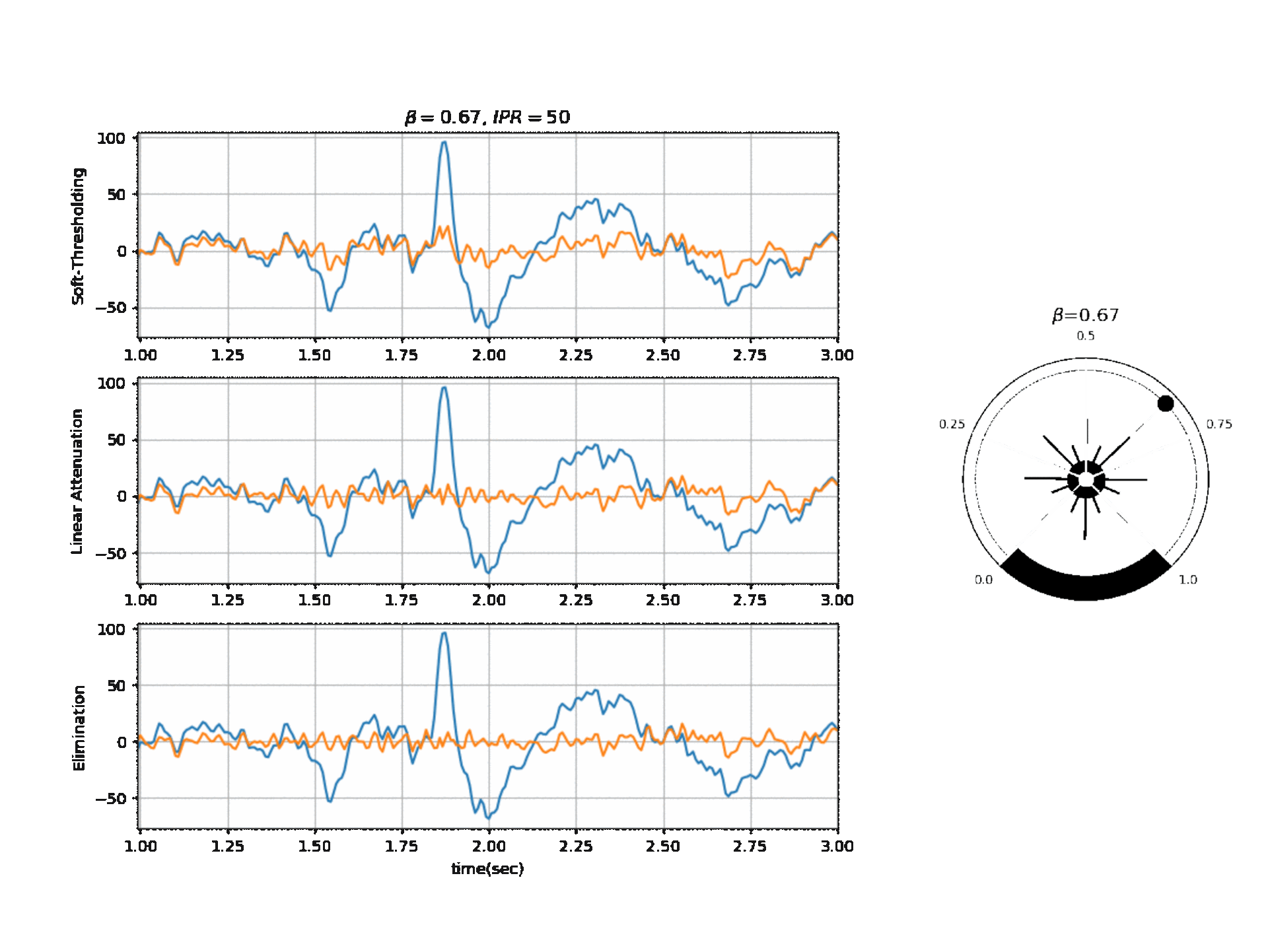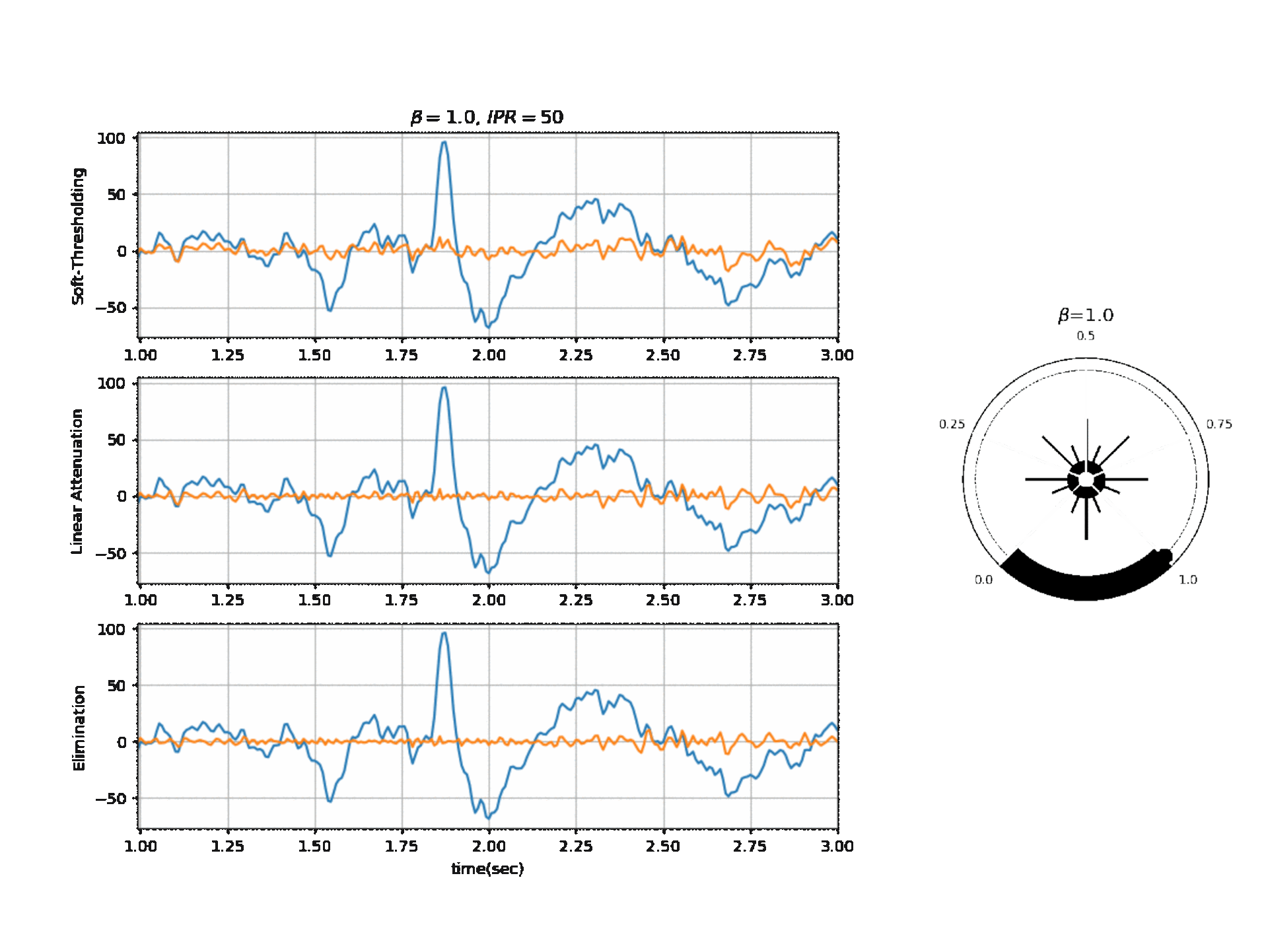 #
#