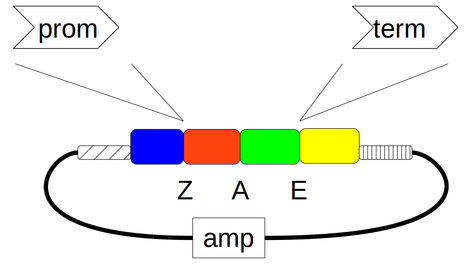
Construction of promoter vector pYPKa_Z_FBA1 and terminator vector pYPKa_E_FBA1¶
This notebook describe the construction of E. coli vectors pYPKa_Z_FBA1 and pYPKa_E_FBA1 with the same insert for which PCR primers are also designed.
The insert defined below is cloned in pYPKa using the blunt restriction enzymes ZraI and EcoRV in two different plasmids. The insert cloned in ZraI will be used as a promoter, while in the EcoRV site the insert will be used as a terminator.
The pydna package is imported in the code cell below. There is a publication describing pydna as well as documentation available online. Pydna is developed on Github.
from pydna.readers import read
from pydna.parsers import parse
from pydna.parsers import parse_primers
from pydna.design import primer_design
from pydna.amplify import pcr
from pydna.amplify import Anneal
The vector backbone pYPKa is read from a local file.
pYPKa = read("pYPKa.gb")
Both restriction enzymes are imported from Biopython
from Bio.Restriction import ZraI, EcoRV
The vector is linearized with both enzymes.
pYPKa_ZraI = pYPKa.linearize(ZraI)
pYPKa_EcoRV = pYPKa.linearize(EcoRV)
The insert sequence is read from a local file. This sequence was parsed from the ypkpathway data file.
ins = read("FBA1.gb")
fp_tail = "ttaaat"
rp_tail = "taattaa"
Primers with the tails above are designed in the code cell below.
ins = primer_design(ins)
fp = fp_tail + ins.forward_primer
rp = rp_tail + ins.reverse_primer
The primers are included in the new_primer.txt list and in the end of the pathway notebook file.
print(fp.format("fasta"))
print(rp.format("fasta"))
with open("new_primers.txt", "a+") as f:
f.write(fp.format("fasta"))
f.write(rp.format("fasta"))
>fw630 FBA1 ttaaatATAACAATACTGACAGTACTAAATAATT >rv630 FBA1 taattaaTTTGAATATGTATTACTTGGTTATG
PCR to create the insert using the newly designed primers.
prd = pcr(fp, rp, ins)
The PCR product has this length in bp.
len(prd)
643
A figure of the primers annealing on template.
prd.figure()
5ATAACAATACTGACAGTACTAAATAATT...CATAACCAAGTAATACATATTCAAA3
||||||||||||||||||||||||| tm 50.7 (dbd) 57.1
3GTATTGGTTCATTATGTATAAGTTTaattaat5
5ttaaatATAACAATACTGACAGTACTAAATAATT3
|||||||||||||||||||||||||||| tm 52.0 (dbd) 55.3
3TATTGTTATGACTGTCATGATTTATTAA...GTATTGGTTCATTATGTATAAGTTT5
A suggested PCR program.
prd.program()
Taq (rate 30 nt/s) 35 cycles |643bp 95.0°C |95.0°C | |Tm formula: Biopython Tm_NN |_________|_____ 72.0°C |72.0°C|SaltC 50mM | 03min00s|30s \ ________|______|Primer1C 1.0µM | | \ 51.5°C/ 0min19s| 5min |Primer2C 1.0µM | | \_____/ | |GC 34% | | 30s | |4-12°C
The final vectors are:
pYPKa_Z_FBA1 = (pYPKa_ZraI + prd).looped().synced(pYPKa)
pYPKa_E_FBA1 = (pYPKa_EcoRV + prd).looped().synced(pYPKa)
The final vectors with reverse inserts are created below. These vectors theoretically make up fifty percent of the clones. The PCR strategy below is used to identify the correct clones.
pYPKa_Z_FBA1b = (pYPKa_ZraI + prd.rc()).looped().synced(pYPKa)
pYPKa_E_FBA1b = (pYPKa_EcoRV + prd.rc()).looped().synced(pYPKa)
A combination of standard primers and the newly designed primers are used for the strategy to identify correct clones. Standard primers are listed here.
p = { x.id: x for x in parse_primers("standard_primers.txt") }
Diagnostic PCR confirmation¶
The correct structure of pYPKa_Z_FBA1 is confirmed by PCR using standard primers 577 and 342 that are vector specific together with the FBA1fw primer specific for the insert in a multiplex PCR reaction with all three primers present.
Two PCR products are expected if the insert was cloned, the sizes depend on the orientation. If the vector is empty or contains another insert, only one product is formed.
Expected PCR products sizes from pYPKa_Z_FBA1:¶
pYPKa_Z_FBA1 with insert in correct orientation.
Anneal( (p['577'], p['342'], fp), pYPKa_Z_FBA1).products
[Amplicon(1577), Amplicon(1409)]
pYPKa_Z_FBA1 with insert in reverse orientation.
Anneal( (p['577'], p['342'], fp), pYPKa_Z_FBA1b).products
[Amplicon(1577), Amplicon(811)]
Empty pYPKa clone.
Anneal( (p['577'], p['342'], fp), pYPKa).products
[Amplicon(934)]
Expected PCR products sizes pYPKa_E_FBA1:¶
pYPKa_E_FBA1 with insert in correct orientation.
Anneal( (p['577'], p['342'], fp), pYPKa_E_FBA1).products
[Amplicon(1577), Amplicon(1328)]
pYPKa_E_FBA1 with insert in reverse orientation.
Anneal( (p['577'], p['342'], fp), pYPKa_E_FBA1b).products
[Amplicon(1577), Amplicon(892)]
The cseguid checksum for the resulting plasmids are calculated for future reference. The cseguid checksum uniquely identifies a circular double stranded sequence.
print(pYPKa_Z_FBA1.cseguid())
print(pYPKa_E_FBA1.cseguid())
OW6W-5Q416AVU9FTAV_I20RdAdU HGisU-gbgw1ephLe7QLqJ0Ghq7c
The sequences are named based on the name of the cloned insert.
pYPKa_Z_FBA1.locus = "pYPKa_Z_FBA1"[:16]
pYPKa_E_FBA1.locus = "pYPKa_Z_FBA1"[:16]
Sequences are stamped with the cseguid checksum. This can be used to verify the integrity of the sequence file.
pYPKa_Z_FBA1.stamp()
pYPKa_E_FBA1.stamp()
cSEGUID_HGisU-gbgw1ephLe7QLqJ0Ghq7c
Sequences are written to local files.
pYPKa_Z_FBA1.write("pYPKa_Z_FBA1.gb")
pYPKa_E_FBA1.write("pYPKa_E_FBA1.gb")
Download pYPKa_Z_FBA1¶
import pydna
reloaded = read("pYPKa_Z_FBA1.gb")
reloaded.verify_stamp()
cSEGUID_OW6W-5Q416AVU9FTAV_I20RdAdU
Download pYPKa_E_FBA1¶
import pydna
reloaded = read("pYPKa_E_FBA1.gb")
reloaded.verify_stamp()
cSEGUID_HGisU-gbgw1ephLe7QLqJ0Ghq7c