Use Case: Unmatched Instances Input¶
Install Dependencies¶
In [1]:
!pip install -r requirements.txt
Requirement already satisfied: panoptica in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from -r requirements.txt (line 1)) (1.0.0.post2.dev0+2f7d01f) Requirement already satisfied: auxiliary in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from -r requirements.txt (line 2)) (0.0.42) Requirement already satisfied: rich in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from -r requirements.txt (line 3)) (13.6.0) Requirement already satisfied: numpy in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from -r requirements.txt (line 4)) (1.25.2) Requirement already satisfied: connected-components-3d<4.0.0,>=3.12.3 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from panoptica->-r requirements.txt (line 1)) (3.12.3) Requirement already satisfied: ruamel.yaml<0.19.0,>=0.18.6 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from panoptica->-r requirements.txt (line 1)) (0.18.6) Requirement already satisfied: scikit-image<0.23.0,>=0.22.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from panoptica->-r requirements.txt (line 1)) (0.22.0) Requirement already satisfied: scipy<2.0.0,>=1.7.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from panoptica->-r requirements.txt (line 1)) (1.11.2) Requirement already satisfied: nibabel>=3.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from auxiliary->-r requirements.txt (line 2)) (5.1.0) Requirement already satisfied: path>=16.10.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from auxiliary->-r requirements.txt (line 2)) (17.0.0) Requirement already satisfied: pathlib>=1.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from auxiliary->-r requirements.txt (line 2)) (1.0.1) Requirement already satisfied: pillow>=10.0.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from auxiliary->-r requirements.txt (line 2)) (10.0.0) Requirement already satisfied: tifffile>=2023.8.25 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from auxiliary->-r requirements.txt (line 2)) (2023.8.30) Requirement already satisfied: markdown-it-py>=2.2.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from rich->-r requirements.txt (line 3)) (3.0.0) Requirement already satisfied: pygments<3.0.0,>=2.13.0 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from rich->-r requirements.txt (line 3)) (2.17.2) Requirement already satisfied: mdurl~=0.1 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from markdown-it-py>=2.2.0->rich->-r requirements.txt (line 3)) (0.1.2) Requirement already satisfied: packaging>=17 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from nibabel>=3.0->auxiliary->-r requirements.txt (line 2)) (23.1) Requirement already satisfied: ruamel.yaml.clib>=0.2.7 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from ruamel.yaml<0.19.0,>=0.18.6->panoptica->-r requirements.txt (line 1)) (0.2.8) Requirement already satisfied: networkx>=2.8 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from scikit-image<0.23.0,>=0.22.0->panoptica->-r requirements.txt (line 1)) (3.1) Requirement already satisfied: imageio>=2.27 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from scikit-image<0.23.0,>=0.22.0->panoptica->-r requirements.txt (line 1)) (2.31.3) Requirement already satisfied: lazy_loader>=0.3 in /opt/anaconda3/envs/seg11panoptdev/lib/python3.11/site-packages (from scikit-image<0.23.0,>=0.22.0->panoptica->-r requirements.txt (line 1)) (0.3)
Setup Imports¶
In [2]:
import numpy as np
from auxiliary.nifti.io import read_nifti
from rich import print as pprint
from panoptica import NaiveThresholdMatching, Panoptica_Evaluator, InputType
from panoptica.utils.segmentation_class import LabelGroup, SegmentationClassGroups
Load Example Data¶
To demonstrate we use a reference and predicition of spine a segmentation with unmatched instances.
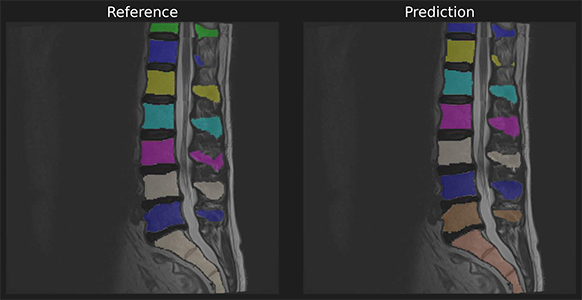
In [3]:
ref_masks = read_nifti("./spine_seg/unmatched_instance/ref.nii.gz")
pred_masks = read_nifti("./spine_seg/unmatched_instance/pred.nii.gz")
# labels are unmatching
pred_masks[pred_masks == 27] = 26 # For later
np.unique(ref_masks), np.unique(pred_masks)
Out[3]:
(array([ 0, 2, 3, 4, 5, 6, 7, 8, 26, 102, 103, 104, 105,
106, 107, 108, 202, 203, 204, 205, 206, 207, 208], dtype=uint8),
array([ 0, 3, 4, 5, 6, 7, 8, 9, 26, 103, 104, 105, 106,
107, 108, 109, 203, 204, 205, 206, 207, 208, 209], dtype=uint8))
Run Evaluation¶
In [4]:
# Define (optionally) semantic groups
# This means that only instance within one group can be matched to each other
segmentation_class_groups = SegmentationClassGroups(
{
"vertebra": LabelGroup(list(range(1, 11))),
"ivd": LabelGroup(list(range(101, 111))),
"sacrum": ([26], True),
"endplate": LabelGroup(list(range(201, 211))),
}
)
# In this case, the label 26 can only be matched with label 26 (thats why have to ensure above that 26 exists in both masks, otherwise they wouldn't be matched)
evaluator = Panoptica_Evaluator(
expected_input=InputType.UNMATCHED_INSTANCE,
instance_matcher=NaiveThresholdMatching(),
# If you want to use segmentation class groups, give it here as argument
segmentation_class_groups=segmentation_class_groups,
)
Inspect Results¶
The results object allows access to individual metrics and provides helper methods for further processing
In [5]:
# print all results
results = evaluator.evaluate(pred_masks, ref_masks, verbose=False)
# The groups will have the names specified above
for groupname, (result, intermediate_steps_data) in results.items():
print()
print("### Group", groupname)
print(result)
────────────────────────────────────────── Thank you for using panoptica ──────────────────────────────────────────
Please support our development by citing
https://github.com/BrainLesion/panoptica#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
### Group vertebra +++ MATCHING +++ Number of instances in reference (num_ref_instances): 7 Number of instances in prediction (num_pred_instances): 7 True Positives (tp): 7 False Positives (fp): 0 False Negatives (fn): 0 Recognition Quality / F1-Score (rq): 1.0 +++ GLOBAL +++ Global Binary Dice (global_bin_dsc): 0.9631786034883428 +++ INSTANCE +++ Segmentation Quality IoU (sq): 0.9259373047661901 +- 0.009654749671578153 Panoptic Quality IoU (pq): 0.9259373047661901 Segmentation Quality Dsc (sq_dsc): 0.9615183012231253 +- 0.005245540988039026 Panoptic Quality Dsc (pq_dsc): 0.9615183012231253 Segmentation Quality ASSD (sq_assd): 0.16832296646947947 +- 0.01828381629759957 Segmentation Quality Relative Volume Difference (sq_rvd): -0.005930868093584259 +- 0.010871203881221219 ### Group ivd +++ MATCHING +++ Number of instances in reference (num_ref_instances): 7 Number of instances in prediction (num_pred_instances): 7 True Positives (tp): 7 False Positives (fp): 0 False Negatives (fn): 0 Recognition Quality / F1-Score (rq): 1.0 +++ GLOBAL +++ Global Binary Dice (global_bin_dsc): 0.9423566613429801 +++ INSTANCE +++ Segmentation Quality IoU (sq): 0.8897861147389462 +- 0.029181150423413706 Panoptic Quality IoU (pq): 0.8897861147389462 Segmentation Quality Dsc (sq_dsc): 0.9414254100052913 +- 0.016436031942319355 Panoptic Quality Dsc (pq_dsc): 0.9414254100052913 Segmentation Quality ASSD (sq_assd): 0.29013503272997326 +- 0.05544330133482135 Segmentation Quality Relative Volume Difference (sq_rvd): 0.020603174193257762 +- 0.03071580120223084 ### Group sacrum +++ MATCHING +++ Number of instances in reference (num_ref_instances): 1 Number of instances in prediction (num_pred_instances): 1 True Positives (tp): 1 False Positives (fp): 0 False Negatives (fn): 0 Recognition Quality / F1-Score (rq): 1.0 +++ GLOBAL +++ Global Binary Dice (global_bin_dsc): 0.9698239455931553 +++ INSTANCE +++ Segmentation Quality IoU (sq): 0.941415733208399 +- 0.0 Panoptic Quality IoU (pq): 0.941415733208399 Segmentation Quality Dsc (sq_dsc): 0.9698239455931553 +- 0.0 Panoptic Quality Dsc (pq_dsc): 0.9698239455931553 Segmentation Quality ASSD (sq_assd): 0.20907172118556794 +- 0.0 Segmentation Quality Relative Volume Difference (sq_rvd): -0.011061174622567414 +- 0.0 ### Group endplate +++ MATCHING +++ Number of instances in reference (num_ref_instances): 7 Number of instances in prediction (num_pred_instances): 7 True Positives (tp): 4 False Positives (fp): 3 False Negatives (fn): 3 Recognition Quality / F1-Score (rq): 0.5714285714285714 +++ GLOBAL +++ Global Binary Dice (global_bin_dsc): 0.6793787581594264 +++ INSTANCE +++ Segmentation Quality IoU (sq): 0.54301762284604 +- 0.01014458743300687 Panoptic Quality IoU (pq): 0.31029578448345146 Segmentation Quality Dsc (sq_dsc): 0.7037824449992637 +- 0.008529812661560601 Panoptic Quality Dsc (pq_dsc): 0.40216139714243637 Segmentation Quality ASSD (sq_assd): 0.33450703853088465 +- 0.010995297631511717 Segmentation Quality Relative Volume Difference (sq_rvd): -0.009548043713894769 +- 0.05397632450411714
In [6]:
# get specific metric, e.g. pq
# Now we need to specify group first
pprint(f"{results['vertebra'][0].pq=}")
results['vertebra'][0].pq=0.9259373047661901
In [7]:
# get dict for further processing, e.g. for pandas
pprint("results dict: ", results["vertebra"][0].to_dict())
results dict: { 'num_ref_instances': 7, 'num_pred_instances': 7, 'tp': 7, 'fp': 0, 'fn': 0, 'prec': 1.0, 'rec': 1.0, 'rq': 1.0, 'sq': 0.9259373047661901, 'sq_std': 0.009654749671578153, 'pq': 0.9259373047661901, 'sq_dsc': 0.9615183012231253, 'sq_dsc_std': 0.005245540988039026, 'pq_dsc': 0.9615183012231253, 'sq_assd': 0.16832296646947947, 'sq_assd_std': 0.01828381629759957, 'sq_rvd': -0.005930868093584259, 'sq_rvd_std': 0.010871203881221219, 'global_bin_dsc': 0.9631786034883428 }
In [9]:
# To inspect different phases, just use the returned intermediate_steps_data object
import numpy as np
for groupname, (result, intermediate_steps_data) in results.items():
print()
print("### Group", groupname)
intermediate_steps_data.original_prediction_arr # yields input prediction array
intermediate_steps_data.original_reference_arr # yields input reference array
# This works with all phases
for i in [InputType.UNMATCHED_INSTANCE, InputType.MATCHED_INSTANCE]:
try:
print(i)
pred = intermediate_steps_data.prediction_arr(i)
ref = intermediate_steps_data.reference_arr(i)
print(
"Prediction array shape =",
pred.shape,
"unique_values=",
np.unique(pred),
)
print(
"Reference array shape =", ref.shape, "unique_values=", np.unique(ref)
)
print()
except AssertionError as e:
print(e)
# This happens because Sacrum class group was set to single_instance, hence the Matching phase is skipped and there is no intermediate result for UNMATCHED_INSTANCE
### Group vertebra InputType.UNMATCHED_INSTANCE Prediction array shape = (164, 399, 17) unique_values= [0 3 4 5 6 7 8 9] Reference array shape = (164, 399, 17) unique_values= [0 2 3 4 5 6 7 8] InputType.MATCHED_INSTANCE Prediction array shape = (164, 399, 17) unique_values= [0 2 3 4 5 6 7 8] Reference array shape = (164, 399, 17) unique_values= [0 2 3 4 5 6 7 8] ### Group ivd InputType.UNMATCHED_INSTANCE Prediction array shape = (96, 406, 17) unique_values= [ 0 103 104 105 106 107 108 109] Reference array shape = (96, 406, 17) unique_values= [ 0 102 103 104 105 106 107 108] InputType.MATCHED_INSTANCE Prediction array shape = (96, 406, 17) unique_values= [ 0 102 103 104 105 106 107 108] Reference array shape = (96, 406, 17) unique_values= [ 0 102 103 104 105 106 107 108] ### Group sacrum InputType.UNMATCHED_INSTANCE key UNMATCHED_INSTANCE not in intermediate steps, maybe the step was skipped? InputType.MATCHED_INSTANCE Prediction array shape = (140, 128, 17) unique_values= [ 0 26] Reference array shape = (140, 128, 17) unique_values= [ 0 26] ### Group endplate InputType.UNMATCHED_INSTANCE Prediction array shape = (85, 385, 17) unique_values= [ 0 203 204 205 206 207 208 209] Reference array shape = (85, 385, 17) unique_values= [ 0 202 203 204 205 206 207 208] InputType.MATCHED_INSTANCE Prediction array shape = (85, 385, 17) unique_values= [ 0 202 204 206 207 209 210 211] Reference array shape = (85, 385, 17) unique_values= [ 0 202 203 204 205 206 207 208]