Warning¶
This tutorial is a work in progress, it does not cover all the functionalities yet. However, it will give you a good overview how you can generate tumor segmentations with BraTS Toolkit.
Glioma segmentation with BraTS Toolkit¶
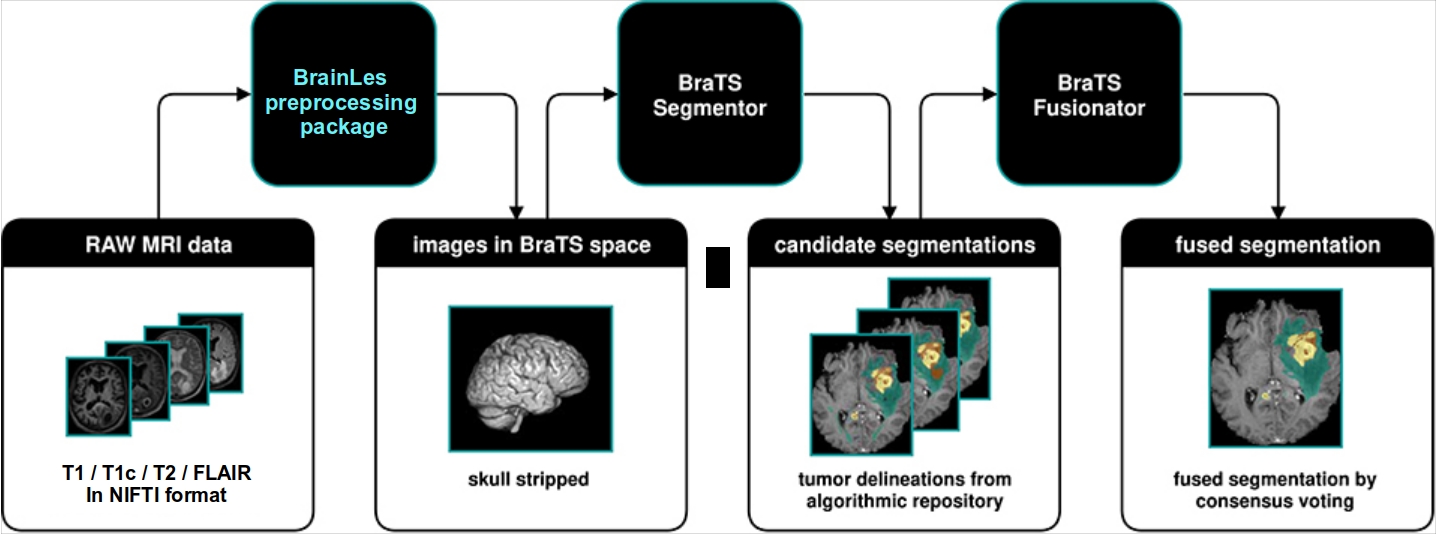
In this Notebook, we will demonstrate how to preprocess brain MR images with the BrainLes preprocessing package. Following we will generate glioma segmentations with multiple algorithms from BraTS Toolkit and fuse them together.
Agenda:¶
Getting started (requirements, installations)
Import of raw data (cMRI)
Preprocessing via BrainLes preprocessig package instead of vanilla preprocessing pipeline from BraTS Toolkit. BraTS challenge algorithms expect preprocessed files (co-registered, skullstripped in SRI-24 space)
Segmentation with BraTS Toolkit
Fusion of generated segementations
1. Getting Started¶
Requirements¶
This tutorial requires:
- Python 3.10+ environment
- Docker (we recommended the most recent version)
optional (but recommended):
- CUDA drivers
- a GPU that is supported (each algorithms supports different set of GPUs)
Run the subsequent cell to check CUDA availability, your python and docker version.
# Your python version (in this notebook) is:
import sys
print(f"Python {sys.version}")
# Check if CUDA is available
import torch
if torch.cuda.is_available():
# Get the number of available CUDA devices
num_cuda_devices = torch.cuda.device_count()
print(
f"CUDA is available with {num_cuda_devices} device(s): {torch.cuda.get_device_name()}"
)
else:
print("CUDA is not available on this system.")
# Print docker version
import subprocess
try:
result = subprocess.run(["docker", "--version"])
except FileNotFoundError as e:
print(
"'docker' seems not to be installed. For installation guide see https://docs.docker.com/get-docker/"
)
Python 3.10.12 (main, Nov 20 2023, 15:14:05) [GCC 11.4.0] CUDA is available with 1 device(s): NVIDIA GeForce GTX 1660 SUPER Docker version 24.0.5, build 24.0.5-0ubuntu1~22.04.1
Installation of dependencies¶
%pip install -U brainles_preprocessing brats_toolkit auxiliary > /dev/null
%pip install -U matplotlib > /dev/null
%load_ext autoreload
%autoreload 2
# package installation may take a while
Note: you may need to restart the kernel to use updated packages. Note: you may need to restart the kernel to use updated packages.
Setup Colab environment (optional)¶
If you installed the packages and requirements on your own machine, you can skip this section and start from the import section. Otherwise you can follow and execute the tutorial on your browser. In order to start working on the notebook, click on the following button, this will open this page in the Colab environment and you will be able to execute the code on your own (Google account required).
(EVA VERSION):
Now that you are visualizing the notebook in Colab, run the next cell to install the packages we will use. There are few things you should follow in order to properly set the notebook up:
- Warning: This notebook was not authored by Google. Click on 'Run anyway'.
- When the installation commands are done, there might be "Restart runtime" button at the end of the output. Please, click it.
If you run the next cell in a Google Colab environment, it will clone the 'tutorials' repository in your google drive. This will create a new folder called "tutorials" in your Google Drive. All generated file will be created/uploaded to your Google Drive respectively.
After the first execution of the next cell, you might receive some warnings and notifications, please follow these instructions:
- 'Permit this notebook to access your Google Drive files?' Click on 'Yes', and select your account.
- Google Drive for desktop wants to access your Google Account. Click on 'Allow'.
This will create the "tutorials" folder. You can navigate it through the lefthand panel in Colab. You might also have received an email that informs you about the access on your Google Drive.
import sys
# Check if we are in google colab currently
try:
import google.colab
colabFlag = True
except ImportError as r:
colabFlag = False
# Execute certain steps only if we are in a colab environment
if colabFlag:
# Create a folder in your Google Drive
from google.colab import drive
drive.mount("/content/drive")
# clone repository and set path
!git clone https://github.com/BrainLesion/tutorials.git /content/drive/MyDrive/tutorials
BASE_PATH = "/content/drive/MyDrive/tutorials/BraTS-Toolkit/"
sys.path.insert(0, BASE_PATH)
else: # normal jupyter notebook environment
BASE_PATH = "./" # current working directory would be BraTs-Toolkit anyways if you are not in colab
2. Import raw input data¶
Raw imput data require
- exam with skull and 4 sequences (T1, T1c, T2, T2-FLAIR), only one file each (in total 4 files)
- file names must end with "t1.nii.gz", "t1c.nii.gz", "t2.nii.gz" and "fla.nii.gz"
- Nifti ".nii.gz" file type
- each exam (4 files) needs to be places in an individual folder within the data folder
<this repository>/BraTS-Toolkit/data/.
The structure is shown using two example exams:
BraTS-Toolkit
├── data
│ ├── TCGA-DU-7294
│ │ ├── *_t1c.nii.gz
│ │ ├── *_t1.nii.gz
│ │ ├── *_t2.nii.gz
│ │ ├── *_fla.nii.gz
│ ├── OtherEXampleFromTCIA
│ │ ├── *_t1c.nii.gz
│ │ ├── *_t1.nii.gz
│ │ ├── *_t2.nii.gz
│ │ ├── *_fla.nii.gz
Example Data¶
Two examples of cMRI from TCIA/TCGA with T1, T1c, T2, T2-FLAIR with skull are provided.
Run the subsequent cell for visualizing one example sample.
from utils import visualize_data, visualize_data2
from pathlib import Path
# paths to example files
t1c_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/MRHR_T1_AX_POST_GAD_OtherEXampleTCIA_TCGA-FG-6692_Si_TCGA-FG-6692_MRHR_T1_AX_POST_GAD_SE_13_se2d1r_t1c.nii.gz"
)
t1_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/T1_AX_OtherEXampleTCIA_TCGA-FG-6692_Si_TCGA-FG-6692_T1_AX_SE_10_se2d1_t1.nii.gz"
)
t2_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/MRHR_T2_AX_OtherEXampleTCIA_TCGA-FG-6692_Si_TCGA-FG-6692_MRHR_T2_AX_SE_2_tse2d1_11_t2.nii.gz"
)
flair_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/MRHR_FLAIR_AX_OtherEXampleTCIA_TCGA-FG-6692_Si_TCGA-FG-6692_MRHR_FLAIR_AX_SE_IR_5_tir2d1_21_fla.nii.gz"
)
# visualize example images
visualize_data2(t1c_path, t1_path, t2_path, flair_path, height_p=0.75)
/home/eva/.local/lib/python3.10/site-packages/matplotlib/projections/__init__.py:63: UserWarning: Unable to import Axes3D. This may be due to multiple versions of Matplotlib being installed (e.g. as a system package and as a pip package). As a result, the 3D projection is not available.
warnings.warn("Unable to import Axes3D. This may be due to multiple versions of "
3. Preprocessing¶
BraTS challenge algorithms expect co-registered, skullstripped files in SRI-24 space, to achieve this preprocessing is required. Instead of using the vanilla preprocessing pipeline from BraTS Toolkit, we recommend using the new BrainLes preprocessing package.
First, we define a function that processes an exam.¶
from auxiliary.normalization.percentile_normalizer import PercentileNormalizer
from auxiliary.turbopath import turbopath
from tqdm import tqdm
from brainles_preprocessing.brain_extraction import HDBetExtractor
from brainles_preprocessing.modality import Modality
from brainles_preprocessing.preprocessor import Preprocessor
from brainles_preprocessing.registration import NiftyRegRegistrator # for linux
# from brainles_preprocessing.registration import ANTsRegistrator # for windows
def preprocess_exam_in_brats_style(inputDir: str) -> None:
"""
Perform BRATS (Brain Tumor Segmentation) style preprocessing on MRI exam data.
Args:
inputDir (str): Path to the directory containing raw MRI files for an exam.
File names must end with "t1.nii.gz", "t1c.nii.gz" "t2.nii.gz" or "fla.nii.gz" to be found!
Raises:
Exception: If any error occurs during the preprocessing.
Example:
brat_style_preprocess_exam("/path/to/exam_directory")
This function preprocesses MRI exam data following the BRATS style, which includes the following steps:
1. Normalization using a percentile normalizer.
2. Registration and correction using NiftyReg.
3. Brain extraction using HDBet.
The processed data is saved in a structured directory within the input directory.
Args:
inputDir (str): Path to the directory containing raw MRI files for an exam.
Returns:
None
"""
inputDir = turbopath(inputDir)
print("*** start ***")
brainles_dir = turbopath(inputDir) + "/" + inputDir.name + "_brainles"
norm_bet_dir = brainles_dir / "normalized_bet"
raw_bet_dir = brainles_dir / "raw_bet"
t1_file = inputDir.files("*t1.nii.gz")
t1c_file = inputDir.files("*t1c.nii.gz")
t2_file = inputDir.files("*t2.nii.gz")
flair_file = inputDir.files("*fla.nii.gz")
# we check that we have only one file of each type
if len(t1_file) == len(t1c_file) == len(t2_file) == len(flair_file) == 1:
t1File = t1_file[0]
t1cFile = t1c_file[0]
t2File = t2_file[0]
flaFile = flair_file[0]
# normalizer
percentile_normalizer = PercentileNormalizer(
lower_percentile=0.1,
upper_percentile=99.9,
lower_limit=0,
upper_limit=1,
)
# define modalities
center = Modality(
modality_name="t1c",
input_path=t1cFile,
raw_bet_output_path=raw_bet_dir / inputDir.name + "_t1c_bet.nii.gz",
atlas_correction=True,
normalizer=percentile_normalizer,
)
moving_modalities = [
Modality(
modality_name="t1",
input_path=t1File,
raw_bet_output_path=raw_bet_dir / inputDir.name + "_t1_bet.nii.gz",
atlas_correction=True,
normalizer=percentile_normalizer,
),
Modality(
modality_name="t2",
input_path=t2File,
raw_bet_output_path=raw_bet_dir / inputDir.name + "_t2_bet.nii.gz",
atlas_correction=True,
normalizer=percentile_normalizer,
),
Modality(
modality_name="flair",
input_path=flaFile,
raw_bet_output_path=raw_bet_dir / inputDir.name + "_fla_bet.nii.gz",
atlas_correction=True,
normalizer=percentile_normalizer,
),
]
preprocessor = Preprocessor(
center_modality=center,
moving_modalities=moving_modalities,
registrator=NiftyRegRegistrator(), # Use this line if you are on linux
# registrator=ANTsRegistrator(), #Use this line if you are on windows
brain_extractor=HDBetExtractor(),
temp_folder="temporary_directory",
limit_cuda_visible_devices="0",
)
preprocessor.run()
/home/eva/.local/lib/python3.10/site-packages/brainles_preprocessing/registration/__init__.py:6: UserWarning: ANTS package not found. If you want to use it, please install it using 'pip install brainles_preprocessing[ants]' warnings.warn(
Now we loop through the exams to preprocess the data:¶
(This may take more than 10 minutes)
EXAMPLE_DATA_DIR = turbopath(f"{BASE_PATH}/data")
exams = EXAMPLE_DATA_DIR.dirs()
for exam in tqdm(exams):
print("processing:", exam)
preprocess_exam_in_brats_style(exam)
0%| | 0/2 [00:00<?, ?it/s]
processing: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294 *** start *** File: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/temporary_directory/atlas-space/atlas__t1c.nii.gz preprocessing... image shape after preprocessing: (103, 160, 160) prediction (CNN id)... 0 1 2 3 4 exporting segmentation...
50%|█████████████████████████████████████████████████████████▌ | 1/2 [06:20<06:20, 380.56s/it]
processing: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA *** start *** File: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/temporary_directory/atlas-space/atlas__t1c.nii.gz preprocessing... image shape after preprocessing: (103, 160, 160) prediction (CNN id)... 0 1 2 3 4 exporting segmentation...
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 2/2 [10:11<00:00, 305.98s/it]
Storage of preprocessed data¶
The preprocessed data is saved in a newly created folder "raw_bet":
data/<datasetName>/<datasetName>_brainles/raw_bet
Run the subsequent cell for visualizing one preprocessed example sample.
from utils import visualize_data2
from pathlib import Path
# paths to example files
t1c_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/raw_bet/OtherEXampleFromTCIA_t1c_bet.nii.gz"
)
t1_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/raw_bet/OtherEXampleFromTCIA_t1_bet.nii.gz"
)
t2_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/raw_bet/OtherEXampleFromTCIA_t2_bet.nii.gz"
)
flair_path = Path(
f"{BASE_PATH}/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/raw_bet/OtherEXampleFromTCIA_fla_bet.nii.gz"
)
# visualize example images
visualize_data2(t1c_path, t1_path, t2_path, flair_path, height_p=0.60)
from brats_toolkit.segmentor import Segmentor
import os
from auxiliary.turbopath import turbopath
from tqdm import tqdm
def segment_exam(
t1_file: str,
t1c_file: str,
t2_file: str,
fla_file: str,
segmentation_file: str,
cid: str,
cuda_device="0",
) -> None:
"""
segment_exam - Segments MRI images using a specified algorithm from the BRATS toolkit.
Parameters:
t1_file (str): Path to the T1-weighted MRI image file.
t1c_file (str): Path to the T1-weighted contrast-enhanced MRI image file.
t2_file (str): Path to the T2-weighted MRI image file.
fla_file (str): Path to the Fluid-attenuated inversion recovery (FLAIR) MRI image file.
segmentation_file (str): Path to the output file where the segmented image will be saved.
cid (str): Algorithm identifier for segmentation.
cuda_device (str, optional): CUDA device ID for GPU acceleration. Default is "0".
Returns:
None
Example:
segment_exam(
t1_file='/path/to/t1.nii.gz',
t1c_file='/path/to/t1c.nii.gz',
t2_file='/path/to/t2.nii.gz',
fla_file='/path/to/fla.nii.gz',
segmentation_file='/path/to/segmentation_result.nii.gz',
cid="mic-dkfz"
)
This function segments MRI images using the specified algorithm from the BRATS toolkit. It accepts paths to T1-weighted,
T1-weighted contrast-enhanced, T2-weighted, and FLAIR MRI images, and performs segmentation using the specified algorithm.
The segmented image is saved in the specified output file.
Note:
- The function uses the BRATS toolkit's Segmentor for segmentation.
- The 'cid' parameter specifies the algorithm to use.
- Segmentation results are saved with a file name corresponding to the algorithm identifier (cid).
- Errors during segmentation are caught, and an error message is printed.
"""
# instantiate
seg = Segmentor(
verbose=True,
gpu=cuda_device,
)
# algorithms we want to select for segmentation
# execute it
if not os.path.exists(segmentation_file):
seg.segment(
t1=t1_file,
t2=t2_file,
t1c=t1c_file,
fla=fla_file,
cid=cid,
outputPath=segmentation_file,
)
else:
print(segmentation_file, "already exists")
Now we again loop through our exams:¶
EXAMPLE_DATA_DIR = turbopath(f"{BASE_PATH}/data")
exams = EXAMPLE_DATA_DIR.dirs()
# we use these five algorithms to segment the images
# 2019 algorithms
# cids = ["mic-dkfz", "scan", "xfeng", "lfb_rwth", "zyx_2019", "scan_2019"]
# 2020 algorithms
cids = ["isen-20", "hnfnetv1-20", "yixinmpl-20", "sanet0-20", "scan-20"]
for exam in tqdm(exams, desc="Exams"):
for cid in tqdm(cids, desc="Algorithms", leave=False):
print("segmenting:", exam, "with algorithm:", cid)
brainles_folder = exam / exam.name + "_brainles"
preprocessed_folder = brainles_folder / "raw_bet"
segmentation_folder = brainles_folder / "segmentation"
segmentation_file = segmentation_folder / cid + ".nii.gz"
segment_exam(
t1_file=preprocessed_folder / exam.name + "_t1_bet.nii.gz",
t1c_file=preprocessed_folder / exam.name + "_t1c_bet.nii.gz",
t2_file=preprocessed_folder / exam.name + "_t2_bet.nii.gz",
fla_file=preprocessed_folder / exam.name + "_fla_bet.nii.gz",
segmentation_file=segmentation_file,
cid=cid,
cuda_device="0",
),
Exams: 0%| | 0/2 [00:00<?, ?it/s] Algorithms: 0%| | 0/5 [00:00<?, ?it/s]
segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294 with algorithm: isen-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/isen-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294 with algorithm: hnfnetv1-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/hnfnetv1-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294 with algorithm: yixinmpl-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/yixinmpl-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294 with algorithm: sanet0-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/sanet0-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294 with algorithm: scan-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/scan-20.nii.gz already exists
Algorithms: 0%| | 0/5 [00:00<?, ?it/s]
segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA with algorithm: isen-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/isen-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA with algorithm: hnfnetv1-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/hnfnetv1-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA with algorithm: yixinmpl-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/yixinmpl-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA with algorithm: sanet0-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/sanet0-20.nii.gz already exists segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA with algorithm: scan-20
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Exams: 100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████| 2/2 [00:00<00:00, 29.71it/s]
/home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/scan-20.nii.gz already exists
The segementations are stored in folder "segmentation"¶
data/<datasetName>/<datasetName>_brainles
Segmentation results are saved with a file name corresponding to the algorithm identifier (cid).
Algorithms used in this notebook:
- yixinmpl-20
- isen-20
- hnfnetv1-20
- scan-20
- sanet0-20
For more details on the algorithms (docker images) see the publication table of the BraTS toolkit
from brats_toolkit.fusionator import Fusionator
def fuse_segmentation_exam(
segmentation_folder: str,
fusion_folder: str,
):
"""
Fuse multiple segmentations using different methods and save the results.
Parameters:
- segmentation_folder (str): Path to the folder containing segmentation files in NIfTI format (*.nii.gz).
- fusion_folder (str): Path to the folder where fused segmentation results will be saved.
Returns:
None
Example:
```python
fuse_segmentation_exam("/path/to/segmentations", "/path/to/fusion_results")
```
This function uses the Fusionator class from the brats_toolkit to fuse segmentations using two different methods:
1. Multimodal Average (MAV) method.
2. Simple fusion method.
The fused segmentations are saved in the specified `fusion_folder` with file names "mav.nii.gz" and "simple.nii.gz".
Note:
- The Fusionator class must be available and correctly imported from the brats_toolkit.
- The segmentation files in the `segmentation_folder` should be in NIfTI format (*.nii.gz).
Args:
- segmentation_folder (str): Path to the folder containing segmentation files in NIfTI format (*.nii.gz).
- fusion_folder (str): Path to the folder where fused segmentation results will be saved.
"""
# instantiate
fus = Fusionator(verbose=True)
# segmentation file paths
segmentations = segmentation_folder.files("*.nii.gz")
# execution
# mav
mavPath = fusion_folder / "mav.nii.gz"
fus.fuse(
segmentations=segmentations,
outputPath=mavPath,
method="mav",
weights=None,
)
# simple
simplePath = fusion_folder / "simple.nii.gz"
fus.fuse(
segmentations=segmentations,
outputPath=simplePath,
method="simple",
weights=None,
)
Now we can again loop through our exams:¶
EXAMPLE_DATA_DIR = turbopath("data")
exams = EXAMPLE_DATA_DIR.dirs()
for exam in tqdm(exams):
print("segmenting:", exam)
brainles_folder = exam / exam.name + "_brainles"
segmentation_folder = brainles_folder / "segmentation"
fusion_folder = brainles_folder / "fusion"
fuse_segmentation_exam(
segmentation_folder=segmentation_folder,
fusion_folder=fusion_folder,
)
0%| | 0/2 [00:00<?, ?it/s]
segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/yixinmpl-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/isen-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/hnfnetv1-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/scan-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/sanet0-20.nii.gz Orchestra: Now fusing all passed .nii.gz files using MAJORITY VOTING. For more output, set the -v or --verbose flag or instantiate the fusionator class with verbose=true Number of segmentations to be fused using compound majority vote is: 5 Labels of current candidate: [0 1 2], dtype: uint8 Labels of current candidate: [0 1 2], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2], dtype: uint8
WARNING:root:No labels passed, choosing those labels automatically: [0 1 2 4]
Labels of current candidate: [0 1 2 4], dtype: uint8 Labels: [1 2 4] weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 5 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 5 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 5 Shape of result: (155, 240, 240) Labels and datatype of result: 2.0 0.0 float64 Converting float64 to uint8 np.ndarray Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/yixinmpl-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/isen-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/hnfnetv1-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/scan-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/TCGA-DU-7294/TCGA-DU-7294_brainles/segmentation/sanet0-20.nii.gz Orchestra: Now fusing all passed .nii.gz files in using SIMPLE. For more output, set the -v or --verbose flag or instantiate the fusionator class with verbose=true Number of segmentations to be fused using SIMPLE is: 5 Labels of current candidate: [0 1 2], dtype: uint8 Labels of current candidate: [0 1 2], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2], dtype: uint8
WARNING:root:No labels passed, choosing those labels automatically: [0 1 2 4]
Labels of current candidate: [0 1 2 4], dtype: uint8 Currently fusing label 1 (155, 240, 240) Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 9122 Candidate with shape (155, 240, 240) and values [0 1] and sum 9180 Candidate with shape (155, 240, 240) and values [0 1] and sum 10541 Candidate with shape (155, 240, 240) and values [0 1] and sum 9115 Candidate with shape (155, 240, 240) and values [0 1] and sum 9608 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 9122 Candidate with shape (155, 240, 240) and values [0 1] and sum 9180 Candidate with shape (155, 240, 240) and values [0 1] and sum 10541 Candidate with shape (155, 240, 240) and values [0 1] and sum 9115 Candidate with shape (155, 240, 240) and values [0 1] and sum 9608 weight is: 3.740271506862001 weight is: 3.8240892486370663 weight is: 3.637696953801965 weight is: 3.69218021577528 weight is: 3.8234751640007185 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Convergence for label 1 after 0 iterations reached. Currently fusing label 2 (155, 240, 240) Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 14547 Candidate with shape (155, 240, 240) and values [0 1] and sum 14527 Candidate with shape (155, 240, 240) and values [0 1] and sum 14450 Candidate with shape (155, 240, 240) and values [0 1] and sum 15764 Candidate with shape (155, 240, 240) and values [0 1] and sum 14593 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 14547 Candidate with shape (155, 240, 240) and values [0 1] and sum 14527 Candidate with shape (155, 240, 240) and values [0 1] and sum 14450 Candidate with shape (155, 240, 240) and values [0 1] and sum 15764 Candidate with shape (155, 240, 240) and values [0 1] and sum 14593 weight is: 3.752705851139802 weight is: 3.7886928242593774 weight is: 3.6542352407582173 weight is: 3.7032811775246737 weight is: 3.849843061189314 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Convergence for label 2 after 0 iterations reached. Currently fusing label 4 (155, 240, 240) Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0] and sum 0 Candidate with shape (155, 240, 240) and values [0] and sum 0 Candidate with shape (155, 240, 240) and values [0 1] and sum 352
WARNING:root:The passed segmentation contains labels other than 1 and 0. WARNING:root:The passed segmentation contains labels other than 1 and 0. WARNING:root:The passed segmentation contains labels other than 1 and 0. ERROR:root:Majority Voting in SIMPLE returned an empty array
Candidate with shape (155, 240, 240) and values [0] and sum 0 Candidate with shape (155, 240, 240) and values [0 1] and sum 279 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 0.0 0.0 float64
/home/eva/.local/lib/python3.10/site-packages/brats_toolkit/fusionator.py:556: RuntimeWarning: invalid value encountered in scalar divide FNR = FN / (FN + TP) /home/eva/.local/lib/python3.10/site-packages/brats_toolkit/fusionator.py:557: RuntimeWarning: invalid value encountered in scalar divide TPR = TP / (TP + FN) /home/eva/.local/lib/python3.10/site-packages/brats_toolkit/fusionator.py:565: RuntimeWarning: invalid value encountered in scalar divide score = 2 * TP / (2 * TP + FP + FN)
Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0] and sum 0 Candidate with shape (155, 240, 240) and values [0] and sum 0
WARNING:root:The passed segmentation contains labels other than 1 and 0. WARNING:root:The passed segmentation contains labels other than 1 and 0.
Candidate with shape (155, 240, 240) and values [0 1] and sum 352 Candidate with shape (155, 240, 240) and values [0] and sum 0 Candidate with shape (155, 240, 240) and values [0 1] and sum 279 weight is: 1 weight is: 1 weight is: 1.0
WARNING:root:The passed segmentation contains labels other than 1 and 0. 50%|██████████████████████████████████████████████████████████ | 1/2 [00:06<00:06, 6.36s/it]
weight is: 1 weight is: 1.0 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 0.0 0.0 float64 Convergence for label 4 after 0 iterations reached. Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 2.0 0.0 float64 Converting float64 to uint8 np.ndarray segmenting: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA
──────────────────────────────────────── Thank you for using BraTS Toolkit ────────────────────────────────────────
Please support our development by citing BraTS Toolkit and the papers of the segmentation algorithms you use:
https://github.com/neuronflow/BraTS-Toolkit#citation -- Thank you!
───────────────────────────────────────────────────────────────────────────────────────────────────────────────────
Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/yixinmpl-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/isen-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/hnfnetv1-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/scan-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/sanet0-20.nii.gz Orchestra: Now fusing all passed .nii.gz files using MAJORITY VOTING. For more output, set the -v or --verbose flag or instantiate the fusionator class with verbose=true Number of segmentations to be fused using compound majority vote is: 5 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8
WARNING:root:No labels passed, choosing those labels automatically: [0 1 2 4]
Labels of current candidate: [0 1 2 4], dtype: uint8 Labels: [1 2 4] weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 5 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 5 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 5 Shape of result: (155, 240, 240) Labels and datatype of result: 4.0 0.0 float64 Converting float64 to uint8 np.ndarray Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/yixinmpl-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/isen-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/hnfnetv1-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/scan-20.nii.gz Loaded: /home/eva/Schreibtisch/GitHub_repos/tutorials_branch/BraTS-Toolkit/data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/segmentation/sanet0-20.nii.gz Orchestra: Now fusing all passed .nii.gz files in using SIMPLE. For more output, set the -v or --verbose flag or instantiate the fusionator class with verbose=true Number of segmentations to be fused using SIMPLE is: 5 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8 Labels of current candidate: [0 1 2 4], dtype: uint8
WARNING:root:No labels passed, choosing those labels automatically: [0 1 2 4]
Labels of current candidate: [0 1 2 4], dtype: uint8 Currently fusing label 1 (155, 240, 240) Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 1886 Candidate with shape (155, 240, 240) and values [0 1] and sum 2155 Candidate with shape (155, 240, 240) and values [0 1] and sum 3410 Candidate with shape (155, 240, 240) and values [0 1] and sum 2867 Candidate with shape (155, 240, 240) and values [0 1] and sum 3005 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 1886 Candidate with shape (155, 240, 240) and values [0 1] and sum 2155 Candidate with shape (155, 240, 240) and values [0 1] and sum 3410 Candidate with shape (155, 240, 240) and values [0 1] and sum 2867 Candidate with shape (155, 240, 240) and values [0 1] and sum 3005 weight is: 3.1511742446548916 weight is: 3.343718586032138 weight is: 3.258616779885049 weight is: 3.0533377014683207 weight is: 3.5990560996214467 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Convergence for label 1 after 0 iterations reached. Currently fusing label 2 (155, 240, 240) Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 24365 Candidate with shape (155, 240, 240) and values [0 1] and sum 26045 Candidate with shape (155, 240, 240) and values [0 1] and sum 19746 Candidate with shape (155, 240, 240) and values [0 1] and sum 16958 Candidate with shape (155, 240, 240) and values [0 1] and sum 19935 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 24365 Candidate with shape (155, 240, 240) and values [0 1] and sum 26045 Candidate with shape (155, 240, 240) and values [0 1] and sum 19746 Candidate with shape (155, 240, 240) and values [0 1] and sum 16958 Candidate with shape (155, 240, 240) and values [0 1] and sum 19935 weight is: 3.6451807045381535 weight is: 3.5102207691555116 weight is: 3.806188837471846 weight is: 3.4225978325341364 weight is: 3.861889228137042 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Convergence for label 2 after 0 iterations reached. Currently fusing label 4 (155, 240, 240) Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 7628 Candidate with shape (155, 240, 240) and values [0 1] and sum 7605 Candidate with shape (155, 240, 240) and values [0 1] and sum 6569 Candidate with shape (155, 240, 240) and values [0 1] and sum 5909 Candidate with shape (155, 240, 240) and values [0 1] and sum 6808 weight is: 1 weight is: 1 weight is: 1 weight is: 1 weight is: 1 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Number of segmentations to be fused using compound majority vote is: 5 Candidate with shape (155, 240, 240) and values [0 1] and sum 7628 Candidate with shape (155, 240, 240) and values [0 1] and sum 7605 Candidate with shape (155, 240, 240) and values [0 1] and sum 6569 Candidate with shape (155, 240, 240) and values [0 1] and sum 5909 Candidate with shape (155, 240, 240) and values [0 1] and sum 6808 weight is: 3.776505044184724 weight is: 3.7866546517920887
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 2/2 [00:12<00:00, 6.31s/it]
weight is: 3.895390628501695 weight is: 3.7132313583169902 weight is: 3.9573242849181356 Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 1.0 0.0 float64 Convergence for label 4 after 0 iterations reached. Shape of result: (155, 240, 240) Shape of current input array: (155, 240, 240) Labels and datatype of current output: 4.0 0.0 float64 Converting float64 to uint8 np.ndarray
The fused segementation is saved in the folder "fusion"¶
BraTS-Toolkit/data/<datasetName>/<datasetName>_brainles/fusion
Run the subsequent cell for visualizing one example sample (T1c with segementation).
The BraTS-Toolkit segementation comprise the GD-enhancing tumor (label 4), the peritumoral edematous tissue (label 2), and the necrotic tumor core (label 1).
from utils import visualize_segmentation
visualize_segmentation(
modality_file="data//OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/raw_bet/OtherEXampleFromTCIA_t1c_bet.nii.gz",
segmentation_file="data/OtherEXampleFromTCIA/OtherEXampleFromTCIA_brainles/fusion/simple.nii.gz",
slice_p=0.63, # default is 0.5
)