Jupyter Notebook Tutorial
Mary Lauren Benton, Ling Chen, and Laura Colbran
- Contents -
Getting Started
Inside Jupyter
Share your notebook!
Tricks and Tips
Other
- Overview -
Jupyter is designed to be an interactive way of presenting your JUlia, PYThon, or Rcode input right next to the figures, along with explanations and comments. It's a good way to document your code and remember which exact snippet made which plot or ran which test.Jupyter had its origins in the IPython notebook, but by 2014 they'd realized that a lot of the functions they were making didn't actually depend on the language being used. So they turned the language-agnostic bits into the Jupyter Project, and kept IPython for the Python-specific aspects of it.
It runs in your web browser, and you can use it to do all your text editing and coding just like normal. The difference is that it has built-in organization and makes it very easy to keep track of what you're doing. It includes a lot of the features of R Markdown files, and can be converted to many other file types, including exectuable scripts, HTML, and PDF.
- How to Install -
Get Anaconda:
Download or lmod (module load Anaconda3)
Create a new Conda environment:
conda create --name myenv
Activate it:
source activate myenv
Start the Notebook App:
jupyter notebook
You'll land here:
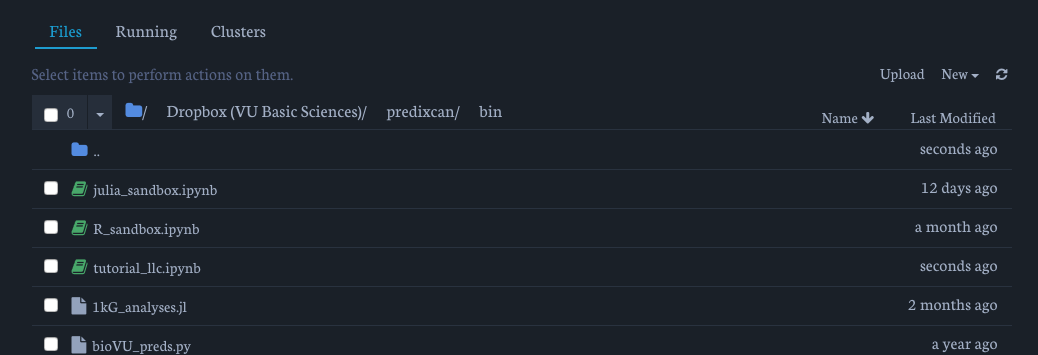
For this example, we'll be using the capra_py3 environment. You can recreate it like this:
conda create --name capra_py3.6 python=3.6
source activate capra_py3.6
conda install rpy2 seaborn jupyterthemes nb_conda
Or, you can use the capra_py3.yml file in the tutorial directory with: conda env create -f capra_py3.yml.
Don't worry too much about understanding this section. We'll write up a specific Conda & virtual environment tutorial.
- Running it via ACCRE -
We can edit these notebooks using the browser on your local machine. We'll launch a job on an ACCRE compute node and then "tunnel" in by connecting our local machine to the node. (Added bonus: you feel like you're doing some legit hacker stuff. But don't worry, it's ACCRE supported.)
Let's look at the template SLURM script:
!cat launch_nb.slurm
With this, all you need to do is (optionally) update the conda environment and the theme command (more on that later!). Sumbit the script using sbatch launch_nb.slurm.
Once your job starts on ACCRE, you can cat notebook.out to see the copy of the ssh command you'll need to use. When I wrote this cell it was:
ssh -L 9999:vmp1015:7777 bentonml@login.accre.vanderbilt.edu
Open a new terminal window and paste that in. You will have to type your ACCRE password. Once you're logged in, you can open your favorit browser window (Chrome, Safari, etc.) and direct it to localhost:9999/. This should bring up Jupyter landing screen you saw above.
Voila! You're now running a Jupyter notebook on the cluster.
- Kernels -
Kernels are basically the back-end that the notebook runs. By default it'll run whatever version of Python was installed along with Anaconda. But what if you work in another language?Use Anaconda to install another 'kernel'.
R:
conda install -c r r-irkernel
Julia:
conda install -c conda-forge julia
Kernels are available for many, many other languages too: Kernels on GitHub
One other note: If you look up in the top corner of your notebook you should see something like this:
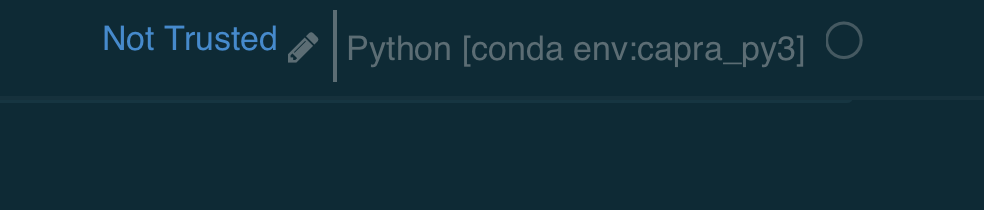
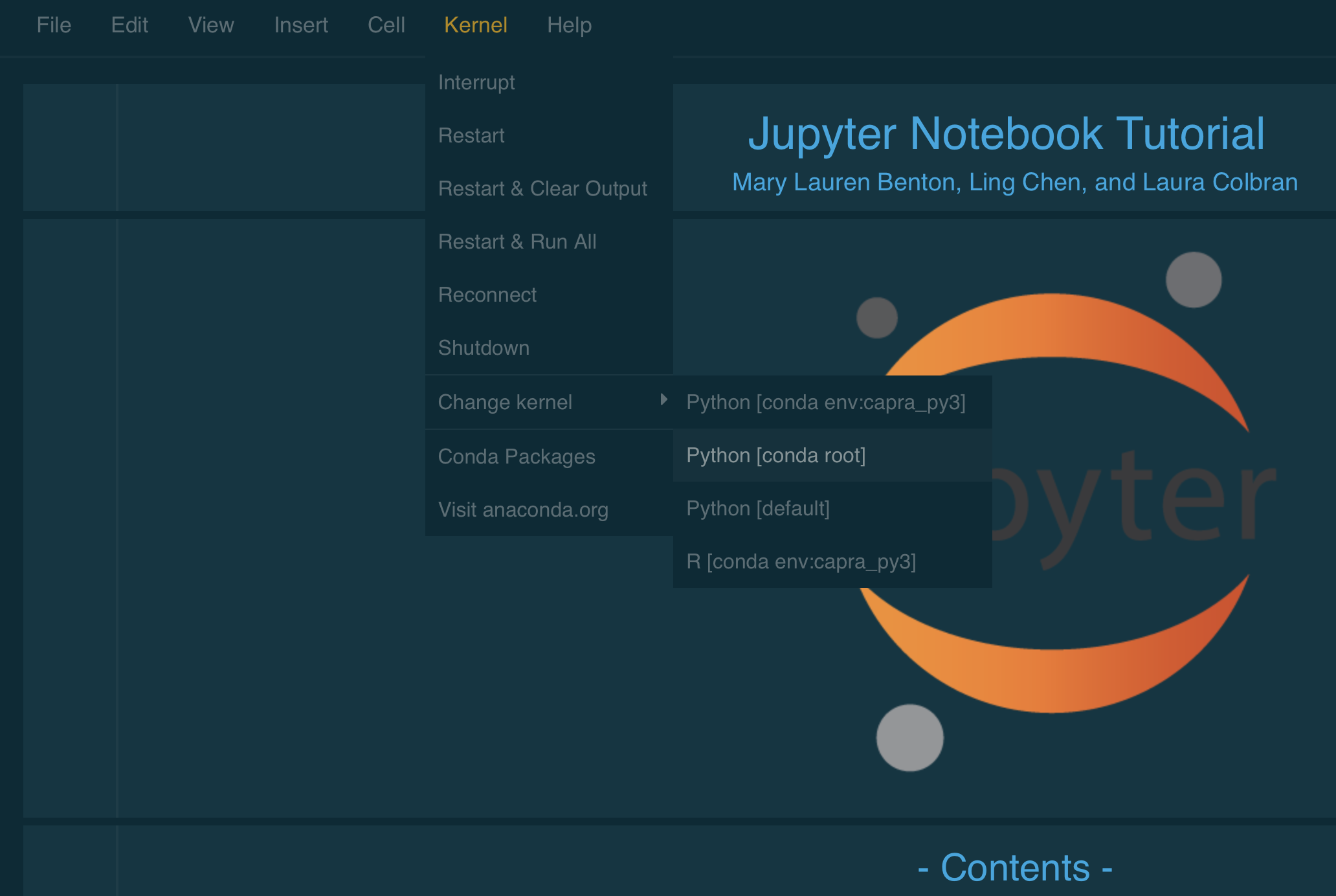
The environment name in [brackets] is the one that is currently running. Double check that this is the environment you intended to run. If not, choose Kernel/Change Kernel from the header menu and pick the one you want.
- Inside Jupyter -
Markdown Cell:
- Normal markdown: Markdown cheat sheet
-
mathjax: a javascript implementation of LaTeX that allows equations to be embedded into HTML.
For example, this markup:$$\log_{2}\frac{\frac{\sum_{1}^{n}Reads_i,mod}{TotalReads_{mod}}}{\frac{\sum_{1}^{n}Reads_i,bg}{TotalReads_{bg}}}$$Turns to: $$\log_{2}\frac{\frac{\sum_{1}^{n}Reads_i,mod}{TotalReads_{mod}}}{\frac{\sum_{1}^{n}Reads_i,bg}{TotalReads_{bg}}}$$
- In a code block,
SympyandLaTexcan be used for math expression too.SymPyis a python library for symbolic mathematics. SymPy documentation.LaTexcan be used with%%latexmagic function:
%%latex
\begin{align}
\LaTeX\\
\nabla \times \vec{\mathbf{B}} -\, \frac1c\, \frac{\partial\vec{\mathbf{E}}}{\partial t} & = \frac{4\pi}{c}\vec{\mathbf{j}} \\
\nabla \cdot \vec{\mathbf{E}} & = 4 \pi \rho \\
\nabla \times \vec{\mathbf{E}}\, +\, \frac1c\, \frac{\partial\vec{\mathbf{B}}}{\partial t} & = \vec{\mathbf{0}} \\
\nabla \cdot \vec{\mathbf{B}} & = 0
\end{align}
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
sns.set()
# Generate an example radial datast
r = np.linspace(0, 10, num=100)
df = pd.DataFrame({'r': r, 'slow': r, 'medium': 2 * r, 'fast': 4 * r})
# Convert the dataframe to long-form or "tidy" format
df = pd.melt(df, id_vars=['r'], var_name='speed', value_name='theta')
# Set up a grid of axes with a polar projection
g = sns.FacetGrid(df, col="speed", hue="speed",
subplot_kws=dict(projection='polar'), size=4.5,
sharex=False, sharey=False, despine=False)
# Draw a scatterplot onto each axes in the grid
g.map(plt.scatter, "theta", "r")
plt.show()
- Magic functions:
There are some predefined magic functions that we can call with a command line style syntax. They all start with%(single line usage) or%%(multiple lines usage).
%lsmagic gives which magic commands we have available in our interpreter.
%lsmagic
%%bash let you run bash commands.
%%bash
bash img/dino.sh danger!
! can be used to run bash commands too.
!bash img/dino.sh danger!
%time, %%time and %timeit, %%timeit are useful to time the execution time of code.
%time x = range(1000)
%%timeit -n 1000
def getx():
x = range(1000)
return x
getx()
%load can be used to load script from file.
%load img/dino2.sh
%load_ext can load an ipython extension by its module name. One useful usage is to load ipython rmagic extension that contains a some magic functions for working with R via rpy2. This extension can be loaded as following:
# if we don't have rpy2 extension, we need to install it first.
# It only supports python3.
#!pip install rpy2
%load_ext rpy2.ipython
%store provides lightweight persistence for python variables.
l = ['hello', 'world']
%store l
exit
l
%store -r
l
%%writefile can write the contents of the cell to a file. The file will be overwritten unless the -a (--append) flag is specified.
You can always see how to use a magic function by typing ? infront of it.
?%%writefile
Raw NBconvert Block:
It can be used for write code that we don't want to run in the notebook, or java script (see hide code trick).- Share your notebook! -
The raw file of Jupyter notebook is a JSON document that contain text, source code, rich media output, and metadata. Each segment of the document is stored in a cell. To render and share it, here are some options.
Convert to other formats with nbconvert:
You can use the nbconvert command to convert the notebook document file to other formats, such as HTML, PDF, LaTex, Markdown, ... For instance, if we want to convert to html file, just do:jupyter nbconvert --to html your_notebook.ipynbPublish notebooks on Github
- Create a repository on github.
- Initiate the current directory with jupyter notebooks as an git repository if not already.
ml Intel git # Lmod load Intel compiler and git git init - Stage and commit the Jupyter notebook you want to push to github.
git add your_notebook.ipynb git commit -m 'some message, such as, your_notebook.ipynb added' - Push to github
git remote add origin your_github_repository_url git push -u origin master - Check it out on github!
Publish as a github gist
Unlike using regular github repository, if you want to share the notebook to others, you have to either make it public or add other people as collaborators. The good thing I like using github gist is that I can create it as a secrete gist that I can share with others. To do this:- Sign in to github
- Navigate to your gist home page.
- Type an optional description and name for your gist.
- Type the text of your gist into the gist text box.
- We can choose to create a public gist or a secret gist.
- Then we can git clone the gist to our local computer and version control, pull, push the gist repository as a regular git repo.
git clone your_gist_url test_gist_notebooks git add your_notebook.ipynb git commit -m 'some message, such as, your_notebook.ipynb added' git remote add origin your_gist_url git push -u origin master - View the notebook on gist, or even better, on nbviewer, just give it the gist ID!
- Tricks and Tips -
Want a clean work report? Hide the code!:
This is one usage of Raw NBconvert cell. Put the following script at the very beginning of the notebook in a Raw NBconvert cell. When the notebook is converted to a html file or viewed through nbviewer website, the code will be hidden. Here is an example jupyter notebook.<script>
function code_toggle() {
if (code_shown){
$('div.input').hide('500');
$('#toggleButton').val('Show Code')
} else {
$('div.input').show('500');
$('#toggleButton').val('Hide Code')
}
code_shown = !code_shown
}
$( document ).ready(function(){
code_shown=false;
$('div.input').hide()
});
</script>
<form action="javascript:code_toggle()"><input type="submit" id="toggleButton" value="Show Code"></form>A pretty theme makes everything better:
I won't write code on a white backgroud. Some people need to choose just the right shade of blue for their headers. We all have quirks. Embrace them with notebook customizations usingjupyterthemes.
We already installed the package into our evironment using conda install jupyterthemes. To activate a specific theme you type jt -t [theme name]. There are also a number of options to customize the font type, font size, line height, margins, cell width, and much more. I like to use the following:
jt -t solarizedd -f anonymous -ofs 10 -cellw 90%
This sets the Solarized Dark theme, with Anonymous Pro as the default font. I increased the cell output font size a bit, and made the default width of my jupyter cells take up 90% of my browser window to give my code a bit of breathing room.
You can apply these themes to the figures within a notebook as well. See the difference:
def rand_uniform(a,b,size=100):
return np.random.rand(size) * (b - a) + a
np.random.seed(90114) # set random seed
x = np.linspace(-1,1)
f = lambda x: np.sin(np.pi * x)
dataX = rand_uniform(-1, 1, 2)
dataY = f(dataX)
best_line = np.poly1d(np.polyfit(dataX, dataY, 1))
plt.plot(x,f(x),label='f(x)');
plt.scatter(dataX, dataY, label='data set', linewidths=1, color='black', facecolor='none', s=200);
plt.plot(x,best_line(x),'--',label='best line');
# import jtplot module in notebook
from jupyterthemes import jtplot
# context == paper, notebook, talk, or poster
jtplot.style(context='notebook', fscale=1.5)
plt.plot(x,f(x),label='f(x)');
plt.scatter(dataX, dataY, label='data set', linewidths=1, color='white', facecolor='none', s=200);
plt.plot(x,best_line(x),'--',label='best line');
More instructions and customizations can be found on the developer's Github here.
Is it Python? Is it R? ...Maybe it's _BOTH_:
Here's an example of some mixed language code using thatrpy2 extension we loaded earlier.
import scipy.stats
# install a quick dependency for this example
#!conda install -y xlrd
# this code is setting up some simulations to recreate Fig 1 from Mostafavi et al. 2017
alpha = 5 * (10**-8)
n_sims = 100
freq_range = np.arange(0.05, 0.5, step=0.05)
# set the potential allele frequency trend
allele_fractions = np.array([1.0, 0.95, 0.9, 0.86, 0.83, 0.81, 0.8, 0.8, 0.81, 0.83, 0.86, 0.9, 0.95, 1.0])
# read in age distribution data from the paper -- note: some nifty pandas tricks are hidden in here!
age_df = pd.read_excel("https://doi.org/10.1371/journal.pbio.2002458.s035", sheet_name="Data for Fig S2")
age_df['APPROX_COUNT'] = round(age_df.FREQ * 57696)
ages = np.array(age_df.APPROX_COUNT, dtype=int)
# set the number of age groups and power values
n_groups = len(ages)
power_py = np.empty(len(freq_range))
power_r = np.empty(len(freq_range))
_ = plt.plot(age_df.AGEBIN.values, allele_fractions, 'o')
%%R
chisquare_r <- function(obs, exp) {
# rescale expected values to match R-- adjusts expected values according to proportion
#prop_exp = exp / sum(exp)
#scaled_exp = sum(obs) * prop_exp
chi <- chisq.test(x = obs, p = exp, rescale = TRUE, correct = FALSE)
return(chi$p.value)
}
for i, freq in enumerate(freq_range):
# set group allele frequencies
group_freqs = allele_fractions * freq
exp_freqs = [freq] * n_groups
# preallocate empty array
sim_p, sim_r = np.empty(n_sims), np.empty(n_sims)
# run x number of simulations at frequency
for sim in range(n_sims):
# calculate observed and expectation
observed = np.random.binomial(n=2*ages, p=group_freqs)
expected = np.random.binomial(n=2*ages, p=exp_freqs)
# rescale expected values to match R-- adjusts expected values according to proportion
prop_exp = expected / sum(expected)
scaled_exp = sum(observed) * prop_exp
# perform chi square in scipy
result = scipy.stats.chisquare(observed, scaled_exp)
sim_p[sim] = result[1]
# perform chi square in r
%R -i observed,expected -o result result=chisquare_r(observed, expected)
sim_r[sim] = result[0]
print("Power (allele frequency = {:.2f}): {:.3f}, {:.3f}".format(freq,
np.sum(sim_p < alpha) / float(n_sims),
np.sum(sim_r < alpha) / float(n_sims)))
- Useful Links -
Additional Information:
Conda CheatsheetR Markdown
Jupyter notebook cheatsheet
Jupyter notebook keyboard shortcuts
Tutorials:
DataCampVandy Bio8366 Ipython and Jupyter tutorial
