Replication: Agosta et al, 2013¶
Introduction¶
This notebook attempts to replicate the following paper with the PPMI dataset:
Agosta, Federica, et al.
The topography of brain damage at different stages of Parkinson's disease., Human brain mapping 34.11 (2013): 2798-2807.
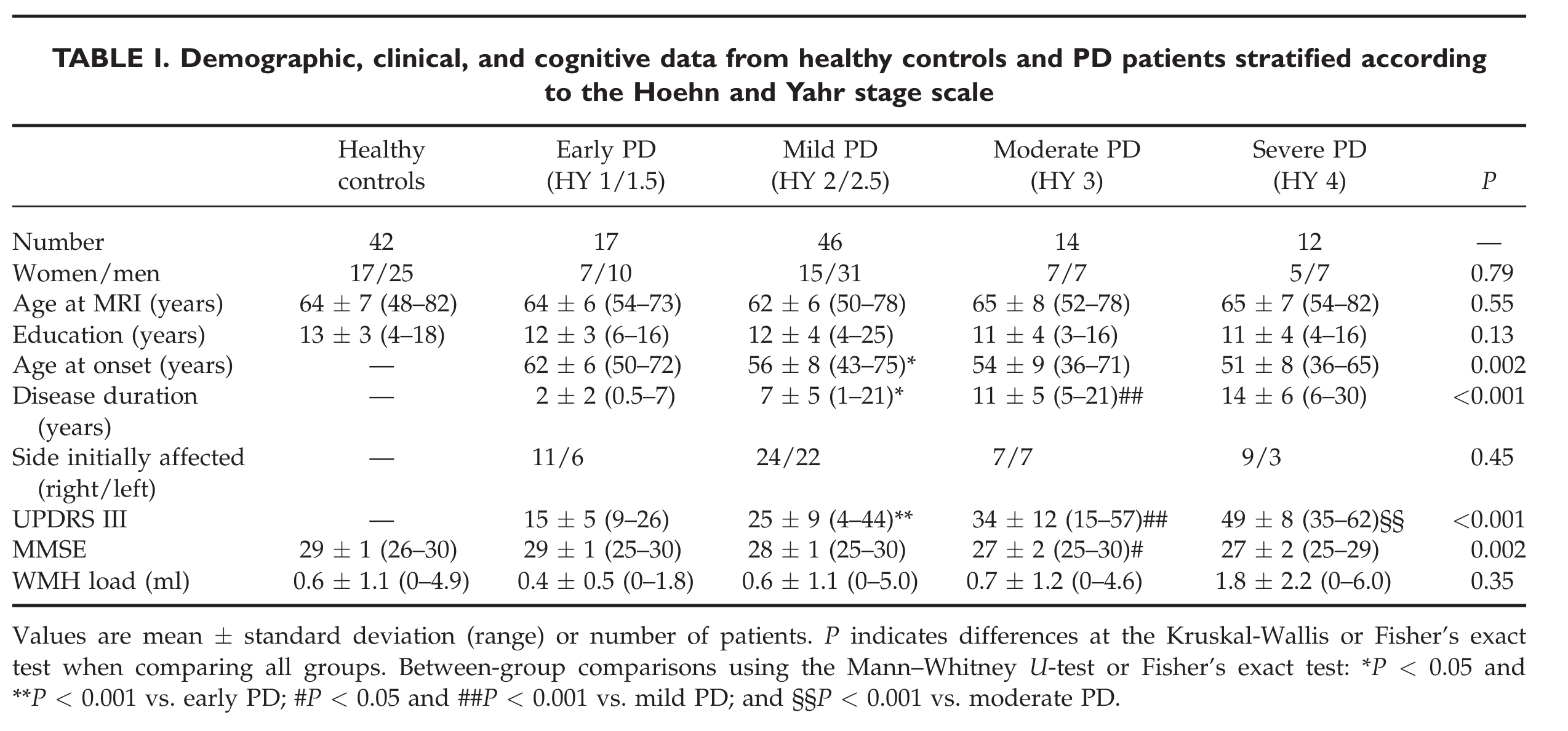
Initial setup¶
In [1]:
import livingpark_utils
utils = livingpark_utils.LivingParkUtils("agosta-etal")
utils.notebook_init()
random_seed = 1
Installing notebook dependencies (see log in install.log)... This notebook was run on 2022-07-21 15:38:37 UTC +0000
PPMI cohort preparation¶
Study data download¶
In [2]:
required_files = [
"iu_genetic_consensus_20220310.csv",
"Cognitive_Categorization.csv",
"Primary_Clinical_Diagnosis.csv",
"Demographics.csv", # SEX
"Socio-Economics.csv", # EDUCYRS
"PD_Diagnosis_History.csv", # Disease duration
"Montreal_Cognitive_Assessment__MoCA_.csv",
"REM_Sleep_Behavior_Disorder_Questionnaire.csv", # STROKE
]
utils.download_ppmi_metadata(required_files)
Download skipped: No missing files!
In [3]:
# H&Y scores
# TODO: move this to livingpark_utils
import os.path as op
file_path = op.join(utils.study_files_dir, "MDS_UPDRS_Part_III_clean.csv")
if not op.exists(file_path):
!(cd {utils.study_files_dir} && python -m wget "https://raw.githubusercontent.com/LivingPark-MRI/ppmi-treatment-and-on-off-status/main/PPMI medication and ON-OFF status.ipynb") # use requests to improve portability
npath = op.join(utils.study_files_dir, "PPMI medication and ON-OFF status.ipynb")
%run "{npath}"
print(f"File {file_path} is now available")
# TODO: move this to livingpark_utils
import os.path as op
file_path = op.join(utils.study_files_dir, "MRI_info.csv")
if not op.exists(file_path):
!(cd {utils.study_files_dir} && python -m wget "https://raw.githubusercontent.com/LivingPark-MRI/ppmi-MRI-metadata/main/MRI metadata.ipynb") # use requests to improve portability
npath = op.join(utils.study_files_dir, "MRI metadata.ipynb")
%run "{npath}"
print(f"File {file_path} is now available")
File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MDS_UPDRS_Part_III_clean.csv is now available File /home/glatard/code/livingpark/agosta-etal/inputs/study_files/MRI_info.csv is now available
Inclusion criteria¶
We obtain the following group sizes:
In [4]:
# H&Y during OFF time
# Exclusion criteria: Patients were excluded if they had:
# (a) parkin, leucine-rich repeat kinase 2 (LRRK2), and glu-
# cocerebrosidase (GBA) gene mutations; OK
# (b) cerebrovascular
# disorders, traumatic brain injury history, or intracranial
# mass; only stroke found in
#
# (c) other major neurological and medical diseases;
# (d) dementia: OK
# DARTEL: 8-mm full width
# at half maximum (FWHM) Gaussian filter
In [5]:
import pandas as pd
# UPDRS3
updrs3 = pd.read_csv(op.join(utils.study_files_dir, "MDS_UPDRS_Part_III_clean.csv"))[
["PATNO", "EVENT_ID", "PDSTATE", "NHY", "NP3TOT"]
]
# Genetics
genetics = pd.read_csv(
op.join(utils.study_files_dir, "iu_genetic_consensus_20220310.csv")
)[["PATNO", "GBA_PATHVAR", "LRRK2_PATHVAR", "NOTES"]]
# Cognitive Categorization
cog_cat = pd.read_csv(op.join(utils.study_files_dir, "Cognitive_Categorization.csv"))[
["PATNO", "EVENT_ID", "COGSTATE"]
]
# Diagnosis
diag = pd.read_csv(op.join(utils.study_files_dir, "Primary_Clinical_Diagnosis.csv"))[
["PATNO", "EVENT_ID", "PRIMDIAG", "OTHNEURO"]
]
# MRI
mri = pd.read_csv(op.join(utils.study_files_dir, "MRI_info.csv"))[
["Subject ID", "Visit code", "Description", "Age"]
]
mri.rename(columns={"Subject ID": "PATNO", "Visit code": "EVENT_ID"}, inplace=True)
# Demographics
demographics = pd.read_csv(op.join(utils.study_files_dir, "Demographics.csv"))[
["PATNO", "SEX"]
]
# Soci-economics
socio_eco = pd.read_csv(op.join(utils.study_files_dir, "Socio-Economics.csv"))[
["PATNO", "EDUCYRS"]
]
# Disease duration
disease_dur = utils.disease_duration()
# MoCA
moca = pd.read_csv(
op.join(utils.study_files_dir, "Montreal_Cognitive_Assessment__MoCA_.csv")
)[["PATNO", "EVENT_ID", "MCATOT"]]
moca["MMSETOT"] = moca["MCATOT"].apply(utils.moca2mmse)
# Stroke
rem = pd.read_csv(
op.join(utils.study_files_dir, "REM_Sleep_Behavior_Disorder_Questionnaire.csv")
)[["PATNO", "EVENT_ID", "STROKE"]]
Download skipped: No missing files!
In [6]:
# Remove genetics data that should be excluded according to PPMI doc
genetics_clean = genetics[
~(
(genetics["NOTES"].notna())
& (
genetics["NOTES"].str.contains("exclude GBA")
| genetics["NOTES"].str.contains("exclude LRRK2")
)
)
]
# Inclusion / exclusion criteria
subjects = (
updrs3[
(updrs3["PDSTATE"] == "OFF") & (updrs3["NHY"].notna()) & (updrs3["NHY"] != "UR")
] # H&Y score is available and was obtained during OFF time
.merge(
genetics_clean[ # No pathogenic GBA or LRRK2 variant present (Note: on-going email thread with Madeleine)
(genetics_clean["GBA_PATHVAR"] == 0)
& (genetics_clean["LRRK2_PATHVAR"] == 0)
],
how="inner",
on="PATNO",
)
.merge(
cog_cat[cog_cat["COGSTATE"] != 3], how="inner", on=["PATNO", "EVENT_ID"]
) # No dementia
.merge(
diag[
(diag["PRIMDIAG"].isin([1, 17])) & (diag["OTHNEURO"].isna())
], # Primary diagnosis is Idiopathic PD or healthy control
how="inner",
on=["PATNO", "EVENT_ID"],
)
.merge(mri, how="inner", on=["PATNO", "EVENT_ID"])
.merge(demographics, how="left", on="PATNO")
.merge(socio_eco, how="left", on="PATNO")
.merge(disease_dur, how="left", on=["PATNO", "EVENT_ID"])
.merge(moca, how="left", on=["PATNO", "EVENT_ID"])
.merge(rem, how="inner", on=["PATNO", "EVENT_ID"])
)
pds = subjects[
(subjects["PRIMDIAG"] == 1) & (subjects["STROKE"] == 0)
] # Include only idiopathic PD with no history of stroke
controls = subjects[
(subjects["PRIMDIAG"] == 17) & (subjects["NHY"] == "0") & (subjects["STROKE"] == 0)
] # Exclude controls with H&Y > 0 and history of stroke
subjects = pd.concat([pds, controls])
print(f"Number of controls: {len(pd.unique(controls['PATNO']))}")
print(f"Number of PD subjects: {len(pd.unique(pds['PATNO']))}")
print(f"Number of PD subject visits by H&Y score:")
pds.groupby(["NHY"], dropna=False).count()[["PATNO"]]
Number of controls: 46 Number of PD subjects: 125 Number of PD subject visits by H&Y score:
Out[6]:
| PATNO | |
|---|---|
| NHY | |
| 1 | 157 |
| 2 | 537 |
| 3 | 34 |
| 4 | 6 |
Cohort matching¶
Matching sex balance as much as possible.
In [7]:
sample_size = {
0: {"total": 42, "women": 17},
1: {"total": 17, "women": 7},
2: {"total": 46, "women": 15},
3: {"total": 14, "women": 7},
4: {"total": 12, "women": 5},
}
samples = {}
subjects_ = subjects.copy() # copy subjects DF to leave it intact
for x in sorted(
sample_size, reverse=True
): # sampling patients in descending H&Y score order as there are less patients with high H&Y scores
print(f"Selecting {sample_size[x]} visits of unique patients with H&Y={x}")
# Sample visits with H&Y=x and make sure they belong to different patients
hy = (
subjects_[subjects_["NHY"] == str(x)]
.groupby(["PATNO"])
.sample(1, random_state=random_seed)
)
# Sample subjects in hy
if len(hy) < sample_size[x]["total"]:
print(
f'Warning: found only {len(hy)} visits of unique patients with H&Y={x} while {sample_size[x]["total"]} were required. '
+ "Including them all regardless of sex balance."
)
samples[x] = hy
else:
# We have enough subjects. Trying to match sex balance
hy_women = hy[hy["SEX"] == 0]
required_women = sample_size[x]["women"]
hy_men = hy[hy["SEX"] == 1]
required_men = sample_size[x]["total"] - required_women
if len(hy_women) < required_women: # We don't have enough women
print(
f"Warning: found only {len(hy_women)} women with H&Y={x} while {required_women} were required: including them all"
)
# We have enough men since we have enough subjects
required_men += required_women - len(hy_women)
required_women = len(hy_women)
if len(hy_men) < required_men:
# We don't have enough men
print(
f"Warning: found only {len(hy_men)} men with H&Y={x} while {required_men} were required: including them all"
)
# We have enough women since we have enough subjects
required_women += required_men - len(hy_men)
required_men = len(hy_men)
samples[x] = pd.concat(
[
hy_women.sample(required_women, random_state=random_seed),
hy_men.sample(required_men, random_state=random_seed),
]
)
# Remove sampled patients from subjects_
subjects_ = subjects_[~(subjects_["PATNO"].isin(samples[x]["PATNO"]))]
Selecting {'total': 12, 'women': 5} visits of unique patients with H&Y=4
Warning: found only 2 visits of unique patients with H&Y=4 while 12 were required. Including them all regardless of sex balance.
Selecting {'total': 14, 'women': 7} visits of unique patients with H&Y=3
Warning: found only 9 visits of unique patients with H&Y=3 while 14 were required. Including them all regardless of sex balance.
Selecting {'total': 46, 'women': 15} visits of unique patients with H&Y=2
Selecting {'total': 17, 'women': 7} visits of unique patients with H&Y=1
Selecting {'total': 42, 'women': 17} visits of unique patients with H&Y=0
Warning: found only 14 women with H&Y=0 while 17 were required: including them all
In [8]:
cohort = pd.concat([samples[x] for x in samples])
In [9]:
def num_stats(column_name, skip_healthy=False):
def round_(x):
if str(x) == "nan":
return float("nan")
else:
return round(x)
a = [
f'{round_(cohort[cohort["NHY"]==str(x)][column_name].mean())} '
+ f'+/- {round_(cohort[cohort["NHY"]==str(x)][column_name].std())} '
+ f'({round_(cohort[cohort["NHY"]==str(x)][column_name].min())}-'
+ f'{round_(cohort[cohort["NHY"]==str(x)][column_name].max())})'
for x in range(5)
]
if skip_healthy:
a[0] = "—"
return a
cohort_stats = pd.DataFrame(
columns=["Healthy controls"] + [f"HY {x}" for x in range(1, 5)]
)
cohort_stats.loc["Number"] = [len(cohort[cohort["NHY"] == str(x)]) for x in range(5)]
cohort_stats.loc["Women / men "] = [
f'{len(cohort[(cohort["NHY"]==str(x)) & (cohort["SEX"]==0)])} / {len(cohort[(cohort["NHY"]==str(x)) & (cohort["SEX"]==1)])}'
for x in range(5)
]
cohort_stats.loc["Age (years)"] = num_stats("Age")
cohort_stats.loc["Education (years)"] = num_stats("EDUCYRS")
cohort_stats.loc["Disease duration (years)"] = num_stats("PDXDUR", skip_healthy=True)
cohort_stats.loc["UPDRS III"] = num_stats("NP3TOT", skip_healthy=True)
cohort_stats.loc["MMSE"] = num_stats("MMSETOT")
In [10]:
cohort_stats
Out[10]:
| Healthy controls | HY 1 | HY 2 | HY 3 | HY 4 | |
|---|---|---|---|---|---|
| Number | 42 | 17 | 46 | 9 | 2 |
| Women / men | 14 / 28 | 7 / 10 | 15 / 31 | 4 / 5 | 1 / 1 |
| Age (years) | 61 +/- 12 (33-83) | 62 +/- 10 (40-79) | 63 +/- 10 (42-78) | 69 +/- 8 (55-80) | 69 +/- 6 (64-73) |
| Education (years) | 16 +/- 3 (12-24) | 14 +/- 3 (8-20) | 15 +/- 3 (9-22) | 14 +/- 3 (8-18) | 20 +/- nan (20-20) |
| Disease duration (years) | — | 4 +/- 3 (0-10) | 4 +/- 3 (0-10) | 4 +/- 2 (0-7) | 2 +/- 2 (1-4) |
| UPDRS III | — | 14 +/- 5 (6-22) | 26 +/- 10 (9-51) | 40 +/- 9 (29-56) | nan +/- nan (nan-nan) |
| MMSE | 30 +/- 1 (28-30) | 29 +/- 1 (26-30) | 29 +/- 2 (22-30) | 28 +/- 2 (26-30) | 29 +/- 0 (29-29) |
Image analysis¶
Structural MRI analysis in the original paper is a straightforward VBM analysis implemented with SPM using the DARTEL toolbox. To replicate it, we mostly followed the excellent tutorial on VBM in SPM available at https://www.fil.ion.ucl.ac.uk/~john/misc/VBMclass15.pdf
Imaging data download¶
Let's first check if the Nifti image files associated with the subjects and visits selected in our cohort are available in cache:
We will now download the missing image files and move them to the data cache:
In [11]:
# utils.download_missing_nifti_files(cohort, link_in_outputs=True)