ATAR: Automatic and Tunable Artifact Removal Algorithm
ATAR: Automatic and Tunable Artifact Removal Algorithm¶
ATAR Algorithm - Automatic and Tunable Artifact Removal Algorithm for EEG Signal.
The algorithm is based on wavelet packet decomposion (WPD), the full description of algorithm can be found here Automatic and Tunable Artifact Removal Algorithm for EEG from the article [1]. Figure 1 shows the the block diagram and operating mode of filtering.
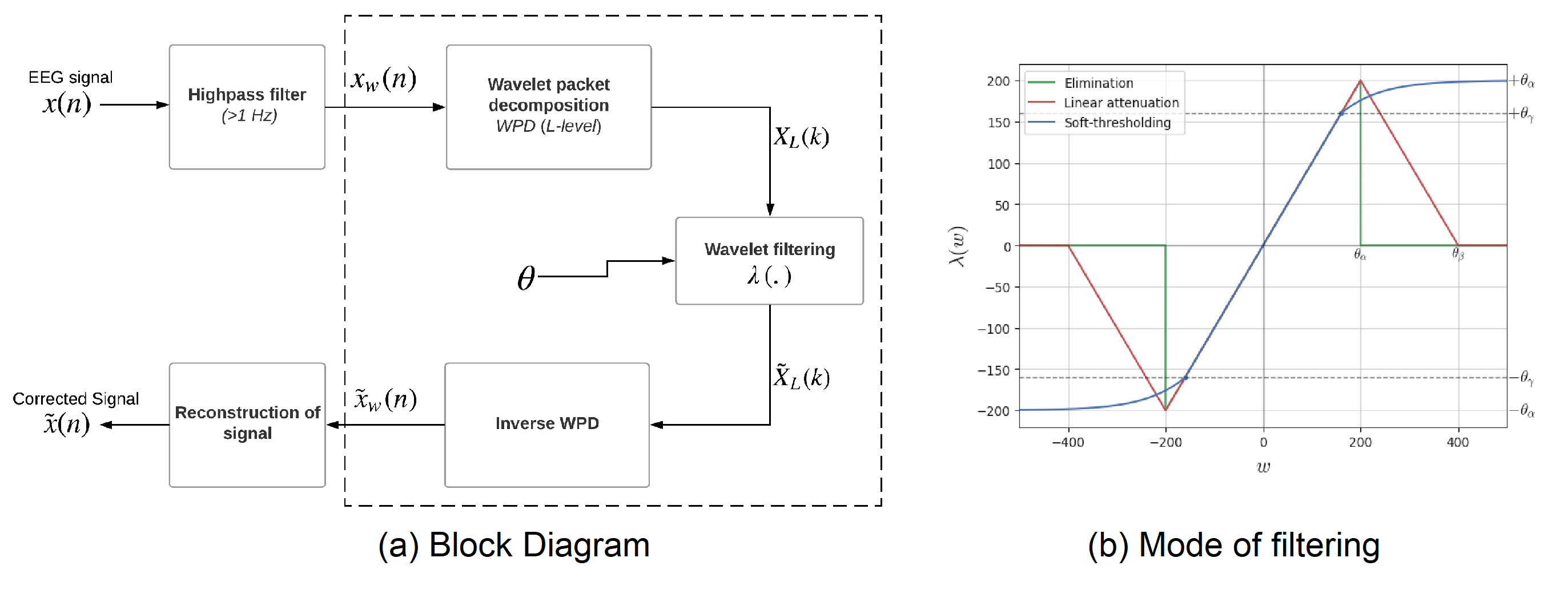
The algorithm is applied on the given multichannel signal X (n,nch), window wise and reconstructed with overall add method. The defualt window size is set to 1 sec (128 samples). For each window, the threshold $\theta_\alpha$ is computed and applied to filter the wavelet coefficients.
There is manily one parameter that can be tuned $\beta$ with different operating modes and other settings. Here is the list of parameters and there simplified meaning given:
Parameters:
$\beta$: This is a main parameter to tune, highher the value, more aggressive the algorithm to remove the artifacts. By default it is set to 0.1. $\beta$ is postive float value.
*OptMode*: This sets the mode of operation, which decides hoe to remove the artifact. By default it is set to 'soft', which means Soft Thresholding, in this mode, rather than removing the pressumed artifact, it is suppressed to the threshold, softly. OptMode='linAtten', suppresses the pressumed artifact depending on how far it is from threshold. Finally, the most common mode - Elimination (OptMode='elim'), which remove the pressumed artifact.
- Soft Thresholding and Linear Attenuation require addition parameters to set the associated thresholds which are by default set to bf=2, gf=0.8.
*wv=db3*: Wavelet funtion, by default set to db3, could be any of ['db3'.....'db38', 'sym2.....sym20', 'coif1.....coif17', 'bior1.1....bior6.8', 'rbio1.1...rbio6.8', 'dmey']
$k_1$, $k_2$: Lower and upper bounds on threshold $\theta_\alpha$.
*IPR=[25,75]*: interpercentile range, range used to compute threshold
Figure 2, below, shows the affect of $\beta$ on a segment of signal with three different modes.
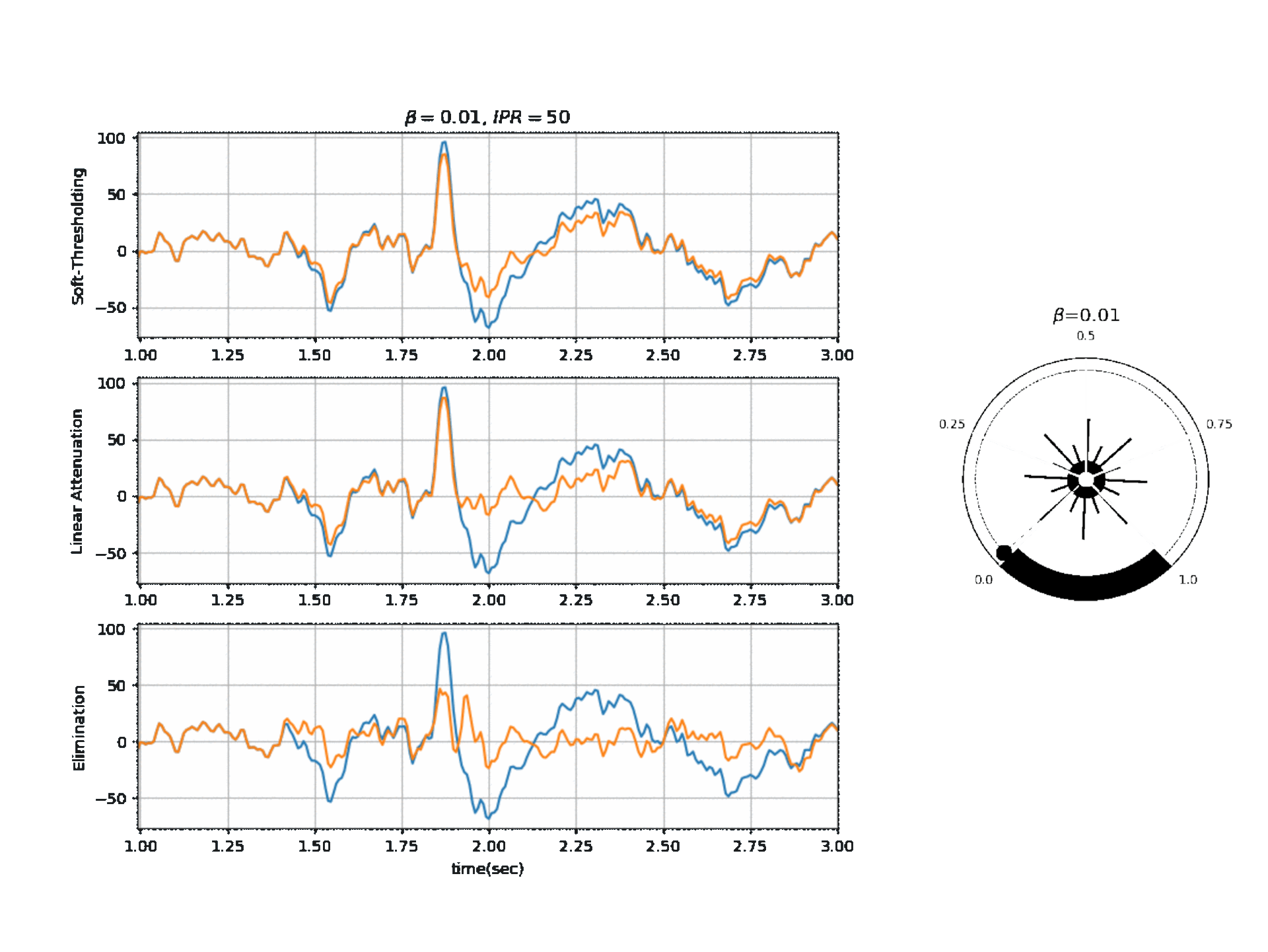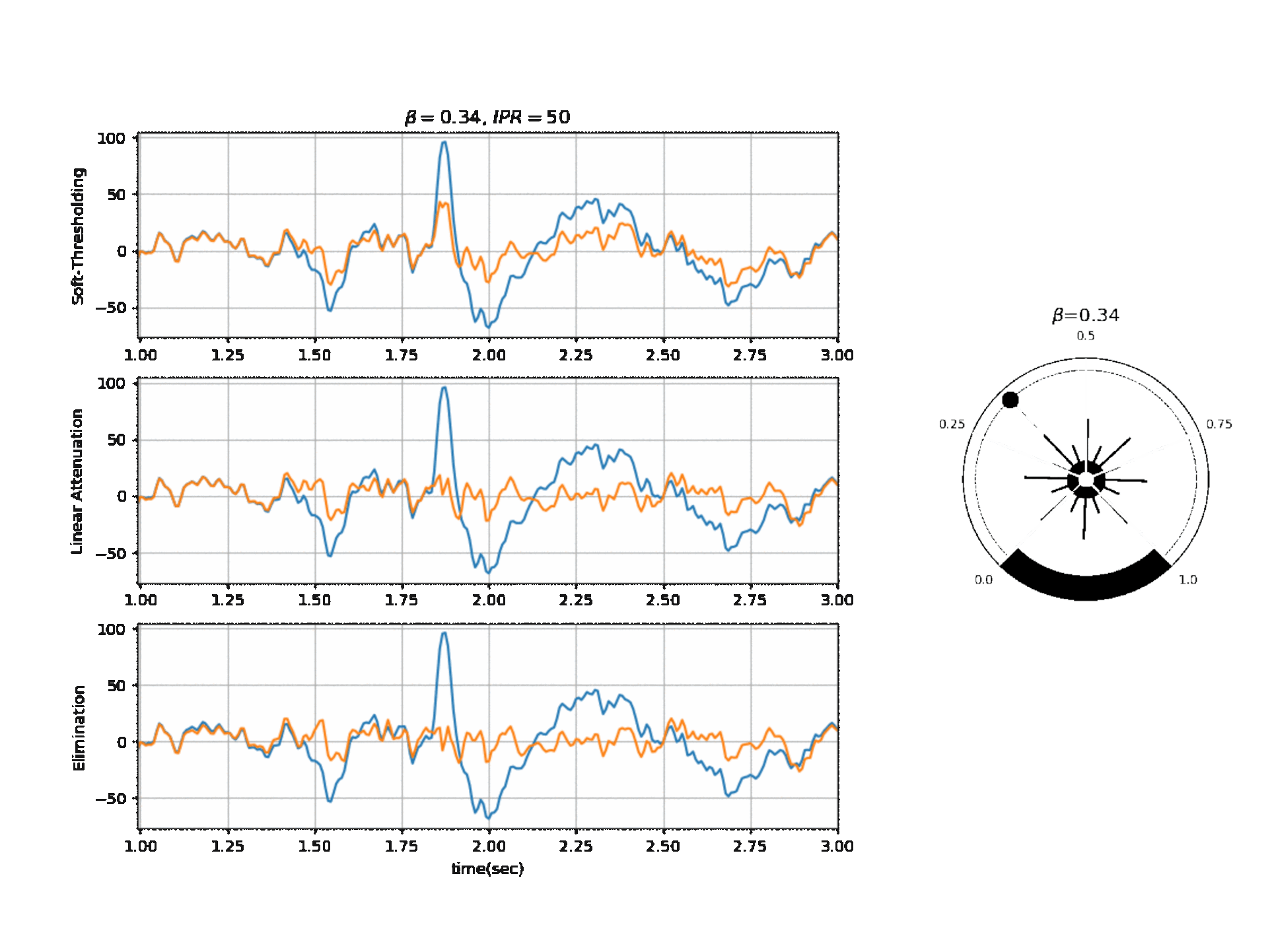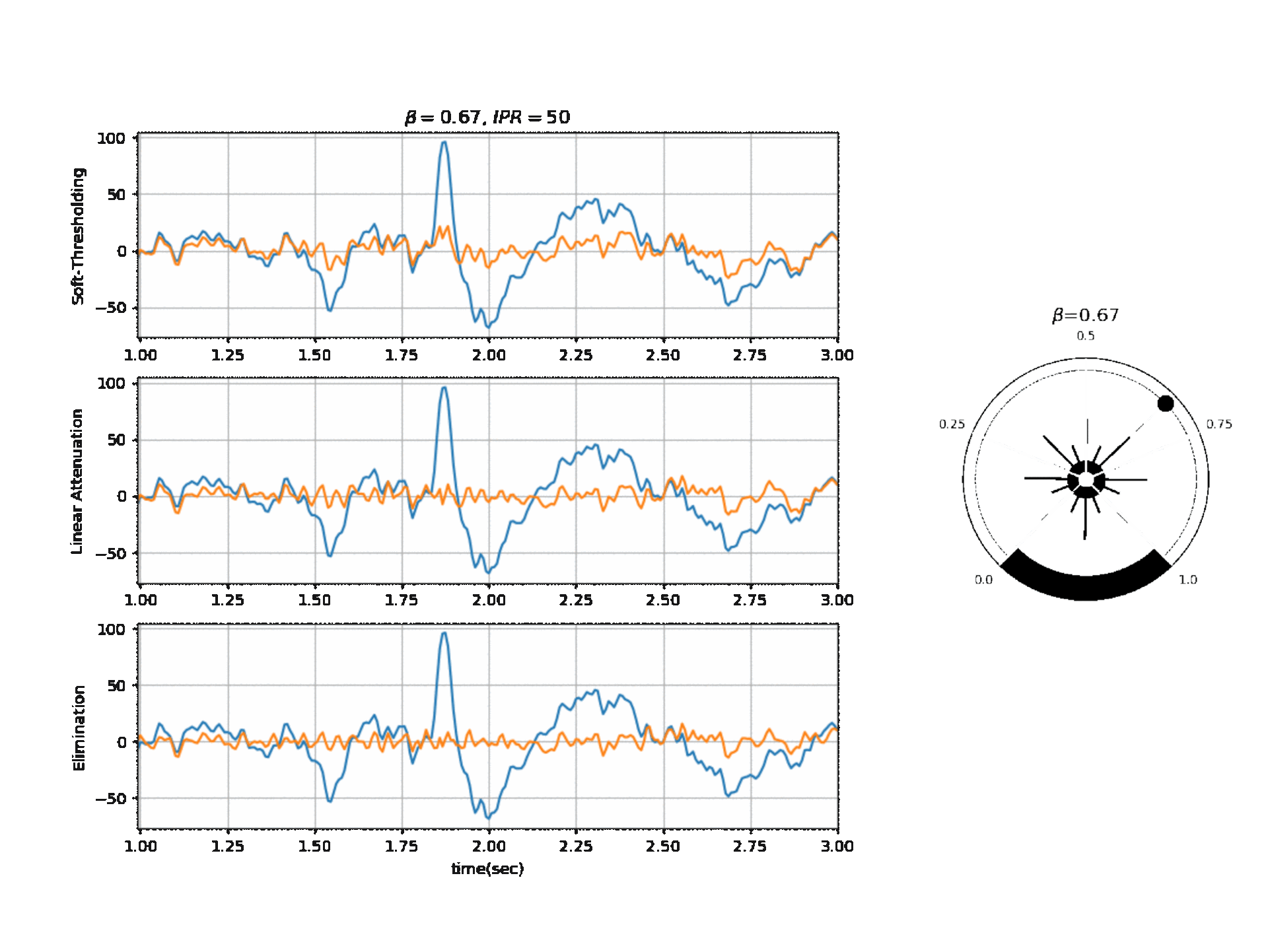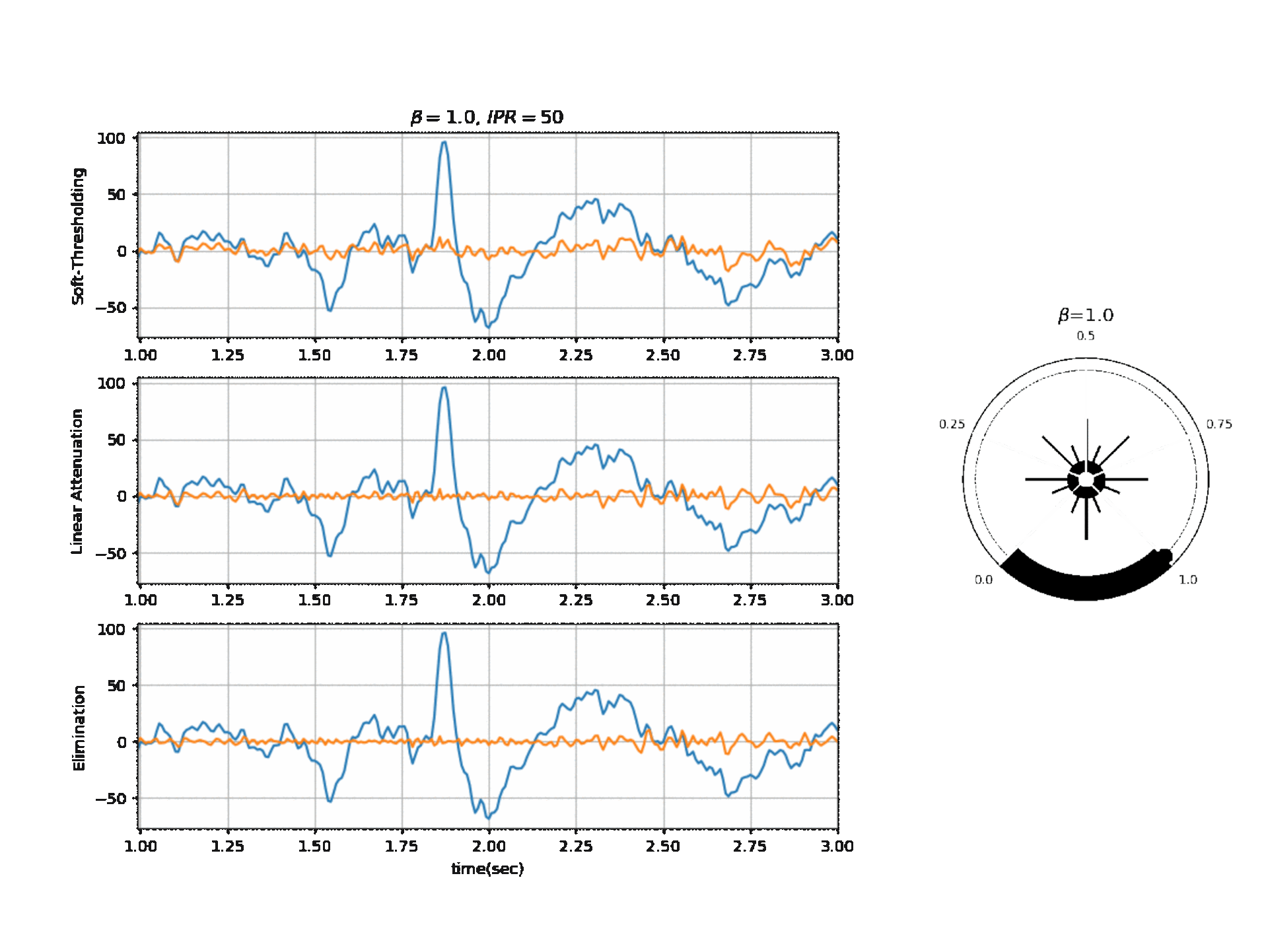
Reference
- [1] Bajaj, Nikesh, et al. "Automatic and tunable algorithm for EEG artifact removal using wavelet decomposition with applications in predictive modeling during auditory tasks." Biomedical Signal Processing and Control 55 (2020): 101624.
There are three functions in spkit.eeg for ATAR algorithm
- spkit.eeg.ATAR(...)
- spkit.eeg.ATAR_1Ch(...)
- spkit.eeg.ATAR_mCh(...)
- spkit.eeg.ATAR_mCh_noParallel(...)
*spkit.eeg.ATAR_1Ch* is for single channel input signal x of shape (n,), where as, *spkit.eeg.ATAR_mCh* is for multichannel signal X with shape (n,ch), which uses joblib for parallel processing of multi channels. For some OS, joblib raise an error of *BrokenProcessPool, in that case use spkit.eeg.ATAR_mCh_noParallel, which is same as spkit.eeg.ATAR_mCh, except parallel processing. Alternatively, use spkit.eeg.ATAR_1Ch* with for loop for each channel.
*spkit.eeg.ATAR* is generalized function, this will call *spkit.eeg.ATAR_1Ch* is single channel is passed else *spkit.eeg.ATAR_mCh* and with use_joblib agrument, it can be set to try parallel processing, else will process each channel individually. We recommed to use *spkit.eeg.ATAR*.
import numpy as np
import matplotlib.pyplot as plt
import spkit as sp
sp.__version__
'0.0.9.4'
Import EEG sample data¶
X,ch_names = sp.load_data.eegSample()
fs = 128
#help(sp.filter_X)
Filter with highpass¶
Xf = sp.filter_X(X,band=[0.5], btype='highpass',fs=fs,verbose=0)
Xf.shape
(2048, 14)
t = np.arange(Xf.shape[0])/fs
plt.figure(figsize=(12,5))
plt.plot(t,Xf+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf: 14 channel - EEG Signal (filtered)')
plt.show()
Applying ATAR Algorithm¶
Soft Thresholding: default settings: OptMode='soft' and $\beta=0.1$¶
XR = sp.eeg.ATAR(Xf.copy(),verbose=0)
XR.shape
(2048, 14)
plt.figure(figsize=(12,5))
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal')
plt.show()
plt.figure(figsize=(12,5))
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Linear Attenuation¶
XR = sp.eeg.ATAR(Xf.copy(),verbose=0,OptMode='linAtten')
XR.shape
(2048, 14)
plt.figure(figsize=(12,5))
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal')
plt.show()
plt.figure(figsize=(12,5))
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Elimination¶
XR = sp.eeg.ATAR(Xf.copy(),verbose=0,OptMode='elim')
XR.shape
(2048, 14)
plt.figure(figsize=(12,5))
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal')
plt.show()
plt.figure(figsize=(12,5))
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Tuning $\beta$ with 'soft' : Controlling the aggressiveness¶
betas = np.r_[np.arange(0.01,0.1,0.02), np.arange(0.1,1, 0.1)].round(2)
for b in betas:
XR = sp.eeg.ATAR(Xf.copy(),verbose=0,beta=b,OptMode='soft')
XR.shape
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$\beta=$' + f'{b}')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Tuning $\beta$ with 'elim'¶
betas = np.r_[np.arange(0.01,0.1,0.02), np.arange(0.1,1, 0.1)].round(2)
for b in betas:
XR = sp.eeg.ATAR(Xf.copy(),verbose=0,beta=b,OptMode='elim')
XR.shape
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$\beta=$' + f'{b}')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
XR = sp.eeg.ATAR(Xf.copy(),wv='db8',beta=0.01,OptMode='elim',verbose=0,)
XR.shape
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$wv=db8$')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
XR = sp.eeg.ATAR_mCh_noParallel(Xf.copy(),wv='db32',beta=0.01,OptMode='elim',verbose=0,)
XR.shape
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$wv=db32$')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Changing upper and lower bounds: $k_1$, $k_2$¶
$k_1$ and $k_2$ are lower and upper bound on the threshold $\theta_\alpha$. $k_1$ is set to 10, which means, the lowest threshold value will be 10, this helps to prevent the removal of entire signal (zeroing out) due to present of high magnitute of artifact. $k_2$ is largest threshold value, which in terms set the decaying curve of threshold $\theta_\alpha$. Increasing k2 will make the removal less aggressive
XR = sp.eeg.ATAR(Xf.copy(),wv='db3',beta=0.1,OptMode='elim',verbose=0,k1=10, k2=200)
XR.shape
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$k_2=200$')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Changing IPR - Interpercentile range¶
IPR is interpercentile range, which is set to 50% (IPR=[25,75]) as default (inter-quartile range), incresing the range increses the aggressiveness of removing artifacts.
XR = sp.eeg.ATAR(Xf.copy(),wv='db3',beta=0.1,OptMode='elim',verbose=0,k1=10, k2=200, IPR=[15,85])
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$IPR=[15,85]$~ 70%')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Using the fix threshold $\theta_\alpha=300$, to all the windows¶
XR = sp.eeg.ATAR(Xf.copy(),wv='db3',thr_method=None,theta_a=300,OptMode='elim',verbose=0)
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$\theta_\alpha=300$')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Changing window length (5 sec)¶
winsize is be default set to 128 (1 sec), assuming 128 sampling rate, which can be changed as needed. In following example it is changed to 5 sec.
XR = sp.eeg.ATAR(Xf.copy(),winsize=128*5,beta=0.01,OptMode='elim',verbose=0,)
plt.figure(figsize=(15,5))
plt.subplot(121)
plt.plot(t,XR+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('XR: Corrected Signal: '+r'$winsize=5sec$')
plt.subplot(122)
plt.plot(t,(Xf-XR)+np.arange(-7,7)*200)
plt.xlim([t[0],t[-1]])
plt.xlabel('time (sec)')
plt.yticks(np.arange(-7,7)*200,ch_names)
plt.grid()
plt.title('Xf - XR: Difference (removed signal)')
plt.show()
Doc¶
help(sp.eeg.ATAR)
Help on function ATAR in module eeg.atar_algorithm:
ATAR(X, wv='db3', winsize=128, thr_method='ipr', IPR=[25, 75], beta=0.1, k1=10, k2=100, est_wmax=100, theta_a=inf, bf=2, gf=0.8, OptMode='soft', wpd_mode='symmetric', wpd_maxlevel=None, factor=1.0, verbose=False, window=['hamming', True], hopesize=None, ReconMethod='custom', packetwise=False, WPD=True, lvl=[], fs=128.0, use_joblib=False)
.
ATAR: - Automatic and Tunable Artifact Removal Algorithm
========================================================
Apply ATAR on short windows of signal (multiple channels - if provided on axis 1:):
Signal is decomposed in smaller overlapping windows and reconstructed after correcting using overlap-add method.
------
for more details, check:
Ref: Bajaj, Nikesh, et al. "Automatic and tunable algorithm for EEG artifact removal using wavelet decomposition with applications in predictive modeling during auditory tasks." Biomedical Signal Processing and Control 55 (2020): 101624.
----------------
input
-----
X: input multi-channel signal of shape (n,ch)
Wavelet family:
wv = ['db3'.....'db38', 'sym2.....sym20', 'coif1.....coif17', 'bior1.1....bior6.8', 'rbio1.1...rbio6.8', 'dmey']
:'db3'(default)
Threshold Computation method:
thr_method : None (default), 'ipr'
: None: fixed threshold theta_a is applied
: ipr : applied with theta_a, bf , gf, beta, k1, k2 and OptMode
: theta_b = bf*theta_a
: theta_g = gf*theta_a
Operating modes:
OptMode = ['soft','elim','linAtten']
: default 'soft'
: use 'elim' with globalgood
Wavelet Decomposition modes:
wpd_mode = ['zero', 'constant', 'symmetric', 'periodic', 'smooth', 'periodization']
default 'symmetric'
Reconstruction Method - Overlap-Add method
ReconMethod : None, 'custom', 'HamWin'
for 'custom': window[0] is used and applied after denoising is window[1] is True else
windowing applied before denoising
output
------
XR: corrected signal of same shape as input X
help(sp.eeg.ATAR_1Ch)
Help on function ATAR_1Ch in module eeg.atar_algorithm:
ATAR_1Ch(x, wv='db3', winsize=128, thr_method='ipr', IPR=[25, 75], beta=0.1, k1=None, k2=100, est_wmax=100, theta_a=inf, bf=2, gf=0.8, OptMode='soft', factor=1.0, wpd_mode='symmetric', wpd_maxlevel=None, verbose=False, window=['hamming', True], hopesize=None, ReconMethod='custom', packetwise=False, WPD=True, lvl=[], fs=128.0)
''
ATAR: - Automatic and Tunable Artifact Removal Algorithm
========================================================
Apply ATAR on short windows of signal (single channel):
Signal is decomposed in smaller overlapping windows and reconstructed after correcting using overlap-add method.
----
for more details, check:
Ref: Bajaj, Nikesh, et al. "Automatic and tunable algorithm for EEG artifact removal using wavelet decomposition with applications in predictive modeling during auditory tasks." Biomedical Signal Processing and Control 55 (2020): 101624.
------------------
Wfilter(x,wv='db3',method=None,IPR=[25,75],beta=0.1,k1=None,k2 =100,
theta_a=np.inf,bf=2,gf=0.8,OptMode ='soft',factor=1.0,showPlot=False,wpd_mode='symmetric',wpd_maxlevel=None)
input
-----
X: input single-channel signal of shape (n,)
Threshold Computation method:
method : None (default), 'ipr'
: provided with theta_a, bf , gf
: theta_b = bf*theta_a
: theta_g = gf*theta_a
Operating modes:
OptMode = ['soft','elim','linAtten']
: default 'soft'
: use 'elim' with global
Wavelet Decomposition modes:
wpd_mode = ['zero', 'constant', 'symmetric', 'periodic', 'smooth', 'periodization']
default 'symmetric'
Wavelet family:
wv = ['db3'.....'db38', 'sym2.....sym20', 'coif1.....coif17', 'bior1.1....bior6.8', 'rbio1.1...rbio6.8', 'dmey']
:'db3'(default)
Reconstruction Methon
ReconMethod : None, 'custom', 'HamWin'
for 'custom': window[0] is used and applied after denoising is window[1] is True else
windowing applied before denoising
output
------
XR: corrected signal of same shape as input X
help(sp.eeg.ATAR_mCh)
Help on function ATAR_mCh in module eeg.atar_algorithm:
ATAR_mCh(X, wv='db3', winsize=128, thr_method='ipr', IPR=[25, 75], beta=0.1, k1=10, k2=100, est_wmax=100, theta_a=inf, bf=2, gf=0.8, OptMode='soft', wpd_mode='symmetric', wpd_maxlevel=None, factor=1.0, verbose=False, window=['hamming', True], hopesize=None, ReconMethod='custom', packetwise=False, WPD=True, lvl=[], fs=128.0, use_joblib=False)
.
ATAR: - Automatic and Tunable Artifact Removal Algorithm
========================================================
Apply ATAR on short windows of signal (multiple channels:):
Signal is decomposed in smaller overlapping windows and reconstructed after correcting using overlap-add method.
------
for more details, check:
Ref: Bajaj, Nikesh, et al. "Automatic and tunable algorithm for EEG artifact removal using wavelet decomposition with applications in predictive modeling during auditory tasks." Biomedical Signal Processing and Control 55 (2020): 101624.
----------------
input
-----
X: input multi-channel signal of shape (n,ch)
Wavelet family:
wv = ['db3'.....'db38', 'sym2.....sym20', 'coif1.....coif17', 'bior1.1....bior6.8', 'rbio1.1...rbio6.8', 'dmey']
:'db3'(default)
Threshold Computation method:
thr_method : None (default), 'ipr'
: None: fixed threshold theta_a is applied
: ipr : applied with theta_a, bf , gf, beta, k1, k2 and OptMode
: theta_b = bf*theta_a
: theta_g = gf*theta_a
Operating modes:
OptMode = ['soft','elim','linAtten']
: default 'soft'
: use 'elim' with globalgood
Wavelet Decomposition modes:
wpd_mode = ['zero', 'constant', 'symmetric', 'periodic', 'smooth', 'periodization']
default 'symmetric'
Reconstruction Method - Overlap-Add method
ReconMethod : None, 'custom', 'HamWin'
for 'custom': window[0] is used and applied after denoising is window[1] is True else
windowing applied before denoising
output
------
XR: corrected signal of same shape as input X