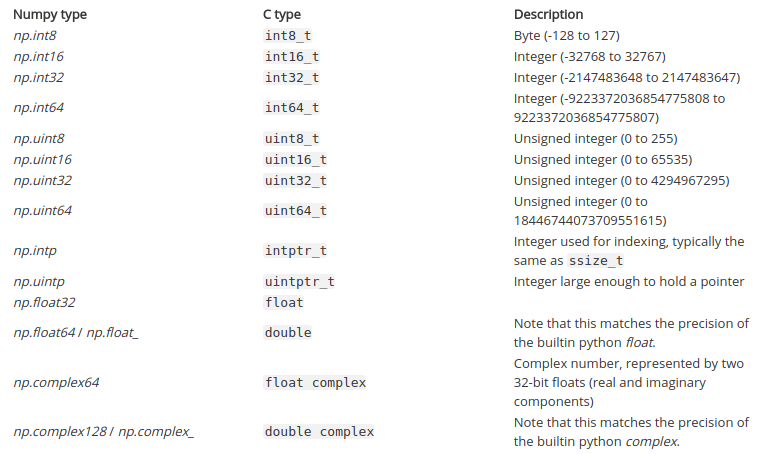
Pandas - Optimize Memory and Speed Operation¶
- Watch Other Interesting Data Science Topics
- Created By: Aakash Goel
- Connect on LinkedIn
- Subscribe on YouTube
- Created on: 17-FEB-2020
- Last Updated on: 06-JUL-2020
Table of contents¶
See Hardware and System Information
1.1. System, CPU, Memory, Swap, Disk, GPU InformationReduce DataFrame size
2.1. Change in int datatype
2.2. Change in float datatype
2.3. Change from object to category datatype
2.4. Convert to Sparse DataFrameReduce DataFrame Loading Time
3.1. Don't Load all columns
3.2. Read Large CSV File (Avoid Memory error and fast reading)Fast Pandas Operation
4.1. Loop and Vectorization
4.2. Indexing
4.3. Parallelization
[1]
 [8]
[8]
1. See Hardware and System Information¶
It is very important to know about your machine on which you are coding.
Hardware and System Information helps you to take proper decision like how much cores you should allocate to your program.
Code for same is taken from here.
1.1 System, CPU, Memory, Swap, Disk, GPU Information¶
# ! pip install tabulate
# ! pip install gputil
import psutil
import platform
import sys
from datetime import datetime
import GPUtil
from tabulate import tabulate
def get_size(bytes, suffix="B"):
"""
Scale bytes to its proper format
e.g:
1253656 => '1.20MB'
1253656678 => '1.17GB'
"""
factor = 1024
for unit in ["", "K", "M", "G", "T", "P"]:
if bytes < factor:
return f"{bytes:.2f}{unit}{suffix}"
bytes /= factor
#### CODE -- https://www.thepythoncode.com/article/get-hardware-system-information-python ####
print("="*40, "Python Version", "="*40)
print (sys.version)
print (sys.version_info)
print("="*40, "System Information", "="*40)
uname = platform.uname()
print(f"System: {uname.system}")
print(f"Release: {uname.release}")
print(f"Version: {uname.version}")
print(f"Machine: {uname.machine}")
print(f"Processor: {uname.processor}")
# let's print CPU information
print("="*40, "CPU Info", "="*40)
# number of cores
print("Physical cores:", psutil.cpu_count(logical=False))
print("Total cores:", psutil.cpu_count(logical=True))
# CPU frequencies
cpufreq = psutil.cpu_freq()
print(f"Max Frequency: {cpufreq.max:.2f}Mhz")
print(f"Min Frequency: {cpufreq.min:.2f}Mhz")
print(f"Current Frequency: {cpufreq.current:.2f}Mhz")
# CPU usage
print("CPU Usage Per Core:")
for i, percentage in enumerate(psutil.cpu_percent(percpu=True, interval=1)):
print(f"Core {i}: {percentage}%")
print(f"Total CPU Usage: {psutil.cpu_percent()}%")
# Memory Information
print("="*40, "Memory Information", "="*40)
# get the memory details
svmem = psutil.virtual_memory()
print(f"Total: {get_size(svmem.total)}")
print(f"Available: {get_size(svmem.available)}")
print(f"Used: {get_size(svmem.used)}")
print(f"Percentage: {svmem.percent}%")
print("="*40, "SWAP", "="*40)
# get the swap memory details (if exists)
swap = psutil.swap_memory()
print(f"Total: {get_size(swap.total)}")
print(f"Free: {get_size(swap.free)}")
print(f"Used: {get_size(swap.used)}")
print(f"Percentage: {swap.percent}%")
# Disk Information
print("="*40, "Disk Information", "="*40)
print("Partitions and Usage:")
# get all disk partitions
partitions = psutil.disk_partitions()
for partition in partitions:
print(f"=== Device: {partition.device} ===")
print(f" Mountpoint: {partition.mountpoint}")
print(f" File system type: {partition.fstype}")
try:
partition_usage = psutil.disk_usage(partition.mountpoint)
except PermissionError:
# this can be catched due to the disk that
# isn't ready
continue
print(f" Total Size: {get_size(partition_usage.total)}")
print(f" Used: {get_size(partition_usage.used)}")
print(f" Free: {get_size(partition_usage.free)}")
print(f" Percentage: {partition_usage.percent}%")
# get IO statistics since boot
disk_io = psutil.disk_io_counters()
print(f"Total read: {get_size(disk_io.read_bytes)}")
print(f"Total write: {get_size(disk_io.write_bytes)}")
# GPU information
print("="*40, "GPU Details", "="*40)
gpus = GPUtil.getGPUs()
list_gpus = []
for gpu in gpus:
# get the GPU id
gpu_id = gpu.id
# name of GPU
gpu_name = gpu.name
# get % percentage of GPU usage of that GPU
gpu_load = f"{gpu.load*100}%"
# get free memory in MB format
gpu_free_memory = f"{gpu.memoryFree}MB"
# get used memory
gpu_used_memory = f"{gpu.memoryUsed}MB"
# get total memory
gpu_total_memory = f"{gpu.memoryTotal}MB"
# get GPU temperature in Celsius
gpu_temperature = f"{gpu.temperature} °C"
gpu_uuid = gpu.uuid
list_gpus.append((
gpu_id, gpu_name, gpu_load, gpu_free_memory, gpu_used_memory,
gpu_total_memory, gpu_temperature, gpu_uuid
))
print(tabulate(list_gpus, headers=("id", "name", "load", "free memory", "used memory", "total memory",
"temperature", "uuid")))
======================================== Python Version ======================================== 3.7.4 (default, Aug 9 2019, 18:34:13) [MSC v.1915 64 bit (AMD64)] sys.version_info(major=3, minor=7, micro=4, releaselevel='final', serial=0) ======================================== System Information ======================================== System: Windows Release: 10 Version: 10.0.18362 Machine: AMD64 Processor: Intel64 Family 6 Model 61 Stepping 4, GenuineIntel ======================================== CPU Info ======================================== Physical cores: 2 Total cores: 4 Max Frequency: 2000.00Mhz Min Frequency: 0.00Mhz Current Frequency: 2000.00Mhz CPU Usage Per Core: Core 0: 56.9% Core 1: 25.0% Core 2: 43.8% Core 3: 42.2% Total CPU Usage: 45.2% ======================================== Memory Information ======================================== Total: 7.91GB Available: 3.85GB Used: 4.06GB Percentage: 51.3% ======================================== SWAP ======================================== Total: 19.26GB Free: 13.41GB Used: 5.85GB Percentage: 30.4% ======================================== Disk Information ======================================== Partitions and Usage: === Device: C:\ === Mountpoint: C:\ File system type: NTFS Total Size: 465.13GB Used: 65.29GB Free: 399.84GB Percentage: 14.0% Total read: 540.46GB Total write: 62.73GB ======================================== GPU Details ======================================== id name load free memory used memory total memory temperature uuid ---- ------ ------ ------------- ------------- -------------- ------------- ------
import numpy as np
import pandas as pd
import nbconvert
from time import time
import gc
import warnings
warnings.filterwarnings("ignore")
2. Reduce DataFrame size¶
2.1 Change in int datatype¶
Situation: Let say, you have Age column having minimum value 1 and maximum value 150, with 10 million total rows in dataframe
Task: Reduce Memory Usage of Age column given above constraints
Action: Change of original dtype from int32 to uint8
Result: Drop from 38.1 MB to 9.5 MB in Memory usage i.e. 75% reduction
## Initializing minimum and maximum value of age
min_age_value , max_age_value = 1,150
## Number of rows in dataframe
nrows = int(np.power(10,7))
## creation of Age dataframe
df_age = pd.DataFrame({'Age':np.random.randint(low=1,high=100,size=nrows)})
## check memory usage before action
df_age.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 10000000 entries, 0 to 9999999 Data columns (total 1 columns): Age int32 dtypes: int32(1) memory usage: 38.1 MB
## Range of "uint8"; satisfies range constraint of Age column
np.iinfo('uint8')
iinfo(min=0, max=255, dtype=uint8)
## Action: conversion of dtype from "int32" to "uint8"
converted_df_age = df_age.astype(np.uint8)
## check memory usage after action
converted_df_age.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 10000000 entries, 0 to 9999999 Data columns (total 1 columns): Age uint8 dtypes: uint8(1) memory usage: 9.5 MB
2.2 Change in float datatype¶
Situation: Let say, you have 50,000 search queries and 5,000 documents and computed cosine similarity for each search query with all documents i.e. dimension 50,000 X 5,000. All similarity values are between 0 and 1 and should have atleast 2 decimal precision
Task: Reduce Memory Usage of cosine smilarity dataframe given above constraints
Action: Change of original dtype from float64 to float16
Result: Drop from 1.9 GB to 476.8 MB or 0.46 GB in Memory usage i.e. 75% reduction
## no. of documents
ncols = int(5*np.power(10,3))
## no. of search queries
nrows = int(5*np.power(10,4))
## creation of cosine similarity dataframe
df_query_doc = pd.DataFrame(np.random.rand(nrows, ncols))
print("No. of search queries: {} and No. of documents: {}".format(df_query_doc.shape[0],df_query_doc.shape[1]))
No. of search queries: 50000 and No. of documents: 5000
## check memory usage before action
df_query_doc.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 50000 entries, 0 to 49999 Columns: 5000 entries, 0 to 4999 dtypes: float64(5000) memory usage: 1.9 GB
## Action: conversion of dtype from "float64" to "float16"
converted_df_query_doc = df_query_doc.astype('float16')
## check memory usage after action
converted_df_query_doc.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 50000 entries, 0 to 49999 Columns: 5000 entries, 0 to 4999 dtypes: float16(5000) memory usage: 476.8 MB
2.3 Change from object to category datatype¶
Situation: Let say, you have Day of Week column having 7 unique values, with 4.9 million total rows in dataframe
Task: Reduce Memory Usage of Day of Week column given only 7 unique value exist
Action: Change of dtype from object to category as ratio of unique values to no. of rows is almost zero
Result: Drop from 2.9 GB to 46.7 MB or 0.045 GB in Memory usage i.e. 98% reduction
## unique values of "days of week"
day_of_week = ["monday","tuesday","wednesday","thursday","friday","saturday","sunday"]
## Number of times day_of_week repeats
repeat_times = 7*np.power(10,6)
## creation of days of week dataframe
df_day_of_week = pd.DataFrame({'day_of_week':np.repeat(a=day_of_week,repeats = repeat_times)})
print("No of rows in days of week dataframe {}".format(df_day_of_week.shape[0]))
No of rows in days of week dataframe 49000000
## check memory usage before action
df_day_of_week.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 49000000 entries, 0 to 48999999 Data columns (total 1 columns): day_of_week object dtypes: object(1) memory usage: 2.9 GB
## Action: conversion of dtype from "object" to "category"
converted_df_day_of_week = df_day_of_week.astype('category')
## check memory usage after action
converted_df_day_of_week.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 49000000 entries, 0 to 48999999 Data columns (total 1 columns): day_of_week category dtypes: category(1) memory usage: 46.7 MB
## check first two rows of dataframe
converted_df_day_of_week.head(2)
| day_of_week | |
|---|---|
| 0 | monday |
| 1 | monday |
## check how mapping of day_of_week is created in category dtype
converted_df_day_of_week.head(2)['day_of_week'].cat.codes
0 1 1 1 dtype: int8
2.4 Convert to Sparse DataFrame¶
Situation: Let say, you have dataframe having large count of zero or missing values (66%) usually happens in lot of NLP task like Count/TF-IDF encoding, Recommender Systems [2]
Task: Reduce Memory Usage of dataframe
Action: Change of DataFrame type to SparseDataFrame as Percentage of Non-Zero Non-NaN values is very less in number
Result: Drop from 228.9 MB to 152.6 MB in Memory usage i.e. 33% reduction
## number of rows in dataframe
nrows = np.power(10,7)
## creation of dataframe
df_dense =pd.DataFrame([[0,0.23,np.nan]]*nrows)
## check memory usage before action
df_dense.info(memory_usage='deep')
<class 'pandas.core.frame.DataFrame'> RangeIndex: 10000000 entries, 0 to 9999999 Data columns (total 3 columns): 0 int64 1 float64 2 float64 dtypes: float64(2), int64(1) memory usage: 228.9 MB
## Percentage of Non-zero and Non-NaN values in dataframe
non_zero_non_nan = np.count_nonzero((df_dense)) - df_dense.isnull().sum().sum()
non_zero_non_nan_percentage = round((non_zero_non_nan/df_dense.size)*100,2)
print("Percentage of Non-Zero Non-NaN values in dataframe {} %".format(non_zero_non_nan_percentage))
Percentage of Non-Zero Non-NaN values in dataframe 33.33 %
## Action: Change of DataFrame type to SparseDataFrame
df_sparse = df_dense.to_sparse()
## check memory usage after action
df_sparse.info(memory_usage='deep')
<class 'pandas.core.sparse.frame.SparseDataFrame'> RangeIndex: 10000000 entries, 0 to 9999999 Data columns (total 3 columns): 0 Sparse[int64, nan] 1 Sparse[float64, nan] 2 Sparse[float64, nan] dtypes: Sparse[float64, nan](2), Sparse[int64, nan](1) memory usage: 152.6 MB
3. Reduce DataFrame Loading Time¶
3.1 Don't Load all columns¶
Situation: Let say, you have csv file which have 100 columns and you only need 10 columns for processing
Task: Reduce pd.read_csv() Loading time
Action: Only Load required columns explicitly i.e. only 10 columns not 100 columns which is default
Result: Drop from 2.8 second to 0.8 second in Loading time i.e. 71% reduction
t0 = time()
ufo = pd.read_csv('http://bit.ly/uforeports')
t1 = time()
print("Default Loading time {} seconds".format(t1-t0))
Default Loading time 2.868640422821045 seconds
t0 = time()
ufo = pd.read_csv('http://bit.ly/uforeports',usecols=['City','State'])
t1 = time()
print("Default Loading time {} seconds".format(t1-t0))
Default Loading time 0.8735167980194092 seconds
3.2 Read Large CSV File¶
Situation: Let say, you have large csv file which is taking too much time in reading or giving memory error while reading in one go
Task: Avoid Memory Error or reduce file loading time
Action 1: Instead of loading file in one go, load file in chunks i.e. use chunksize parameter in read_csv. Useful when you don't need to load all data into memory at once
Action 2: See if you can do memory saving by doing following operations:
- Explicit pass appropriate
dtypesfor each column while loading file and reduce the volume of the data. Example - Sometimes64-bitprecision is overkill and can use32-bit, 2x memory savings are worth it. Usecolsparameter inread_csvi.e. only load required columnsData compression- Here, I don't mean by zip or gzip file. Say your status column has two values: "AVAILABLE" and "UNAVAILABLE". Instead of storing them as string (~ 10 bytes per entry), store as boolean: True or False (1 byte per entry)
Action 3: For faster data loading, serialize data (pickle) on the first run and load the serialized data instead of the csv files in subsequent runs.[15]
Action 4: Use of other Library instead of pandas but with similar functionality:
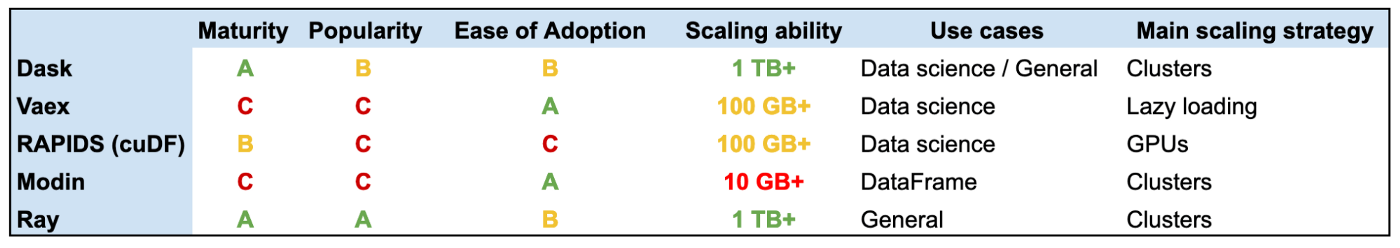
Dask - Dask provides multi-core execution on larger-than-memory datasets.
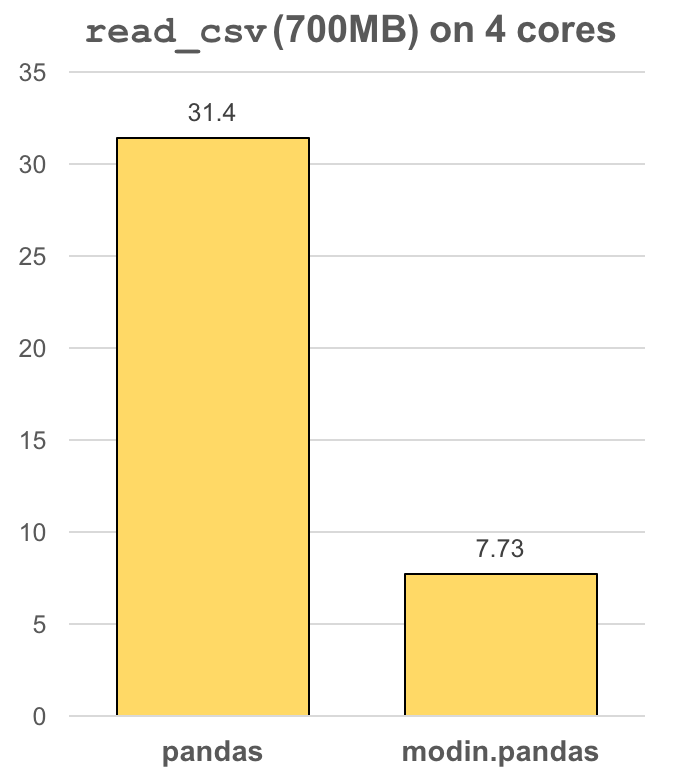
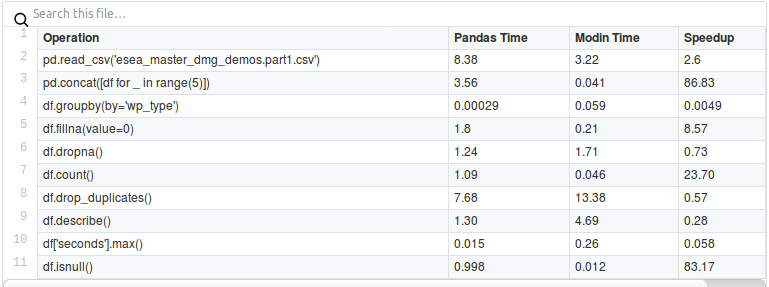
Modin - Modin transparently distributes the data and computation. Modin provides speed-ups of up to 4x on a laptop with 4 physical cores. Pandas able to use one core at a time when you are doing computation of any kind. With Modin, you are able to use all of the CPU cores on your machine.
 [9]
[9]
 [11]
[11]
# # pip install modin[ray]
# import ray
# ray.init(num_cpus=4)
# import modin.pandas as pd
# export MODIN_OUT_OF_CORE=true
 [13]
[13]
4. Fast Pandas Operation¶
4.1 Loop and Vectorization¶
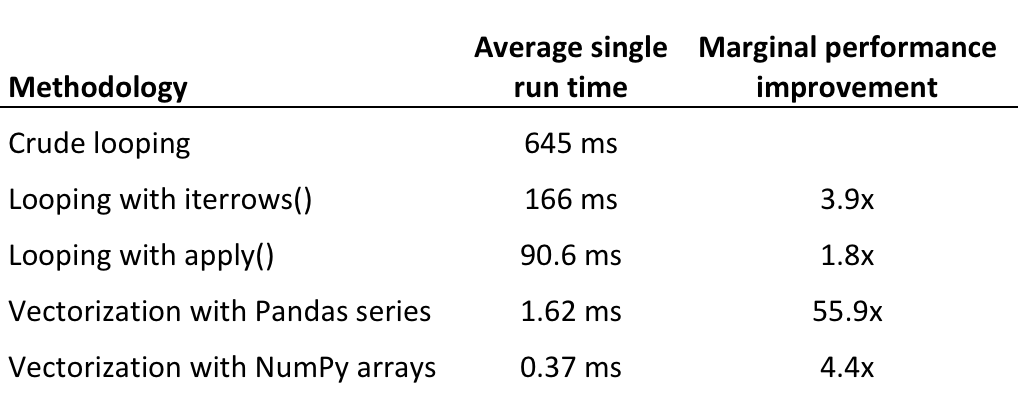 [16]
[16]
 [17]
[17]
4.2 Indexing¶
Situation: Let say, you want to join two dataframes based on some common column
Task: Reduce pd.merge() time
Action: During merge operation, use indexing
Result: Drop from 10 second to 2 second in Loading time i.e. 80% reduction
General Advice: Use indexing as much as possible for merging and value lookup in dataframe [18]
df1 = pd.DataFrame({'A':list(range(10000000)),'B':list(range(0,20000000,2))})
df2 = pd.DataFrame({'A':list(range(10000000)),'C':list(range(0,40000000,4))})
t0 = time()
df = pd.merge(df1,df2,how='left',on='A')
t1 = time()
print("Time Taken in merge before indexing {}".format(t1-t0))
gc.collect()
t0 = time()
df1_ = df1.set_index('A')
df2_ = df2.set_index('A')
df_ = pd.merge(df1_,df2_, left_index=True, right_index=True)
t1 = time()
print("Time Taken in merge after indexing {}".format(t1-t0))
Time Taken in merge before indexing 9.495047569274902 Time Taken in merge after indexing 2.741480827331543
4.3 Parallelization¶
Question: Can we parallelize pandas apply or groupby function without much change in code ?
Answer: Yes, there are lot of libraries which can help in parallelizing pandas apply function without much change in code.
Libraries
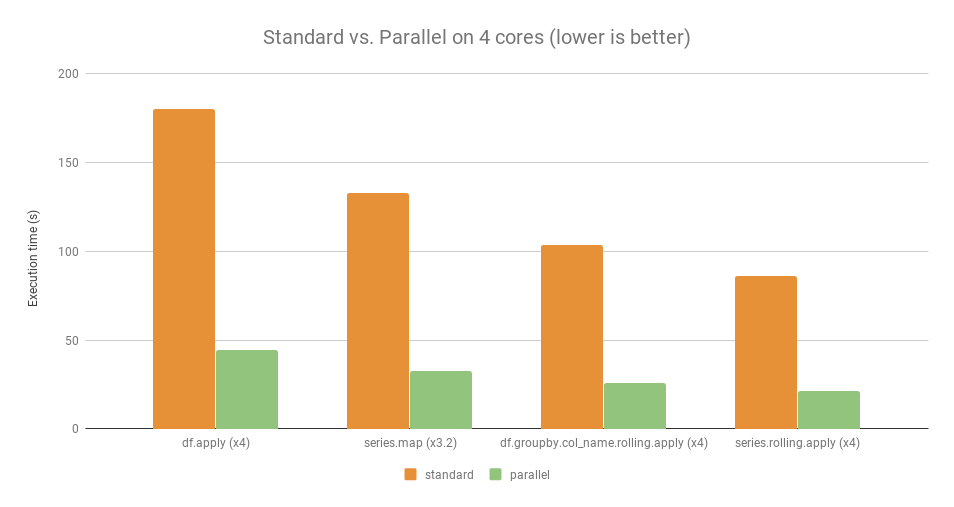 [19]
[19]
General Advice: Parallelization is efficient only if the amount of calculation to parallelize is high enough. For very little amount of data, using parallelization is not always worth it.
#########################################
#### PANDARALLEL CODE #################
#########################################
# from pandarallel import pandarallel
# pandarallel.initialize(nb_workers = _NO_CORES_USED_FOR_PARALLELIZATION,progress_bar = True)
# df.parallel_apply(func)
# df.groupby(args).parallel_apply(func)
#########################################
#### SWIFTER CODE #################
#########################################
# import pandas as pd
# import swifter
# df['Col'].swifter.apply(anyfunction)
5. References¶
https://machinelearningmastery.com/sparse-matrices-for-machine-learning/
https://stackoverflow.com/questions/39100971/how-do-i-release-memory-used-by-a-pandas-dataframe
https://pandas.pydata.org/pandas-docs/stable/user_guide/sparse.html
https://cmdlinetips.com/2018/03/sparse-matrices-in-python-with-scipy/
https://jakevdp.github.io/blog/2014/05/09/why-python-is-slow/
https://towardsdatascience.com/scaling-pandas-comparing-dask-ray-modin-vaex-and-rapids-c74c85a4e59c
Modin Vs Pandas https://www.kdnuggets.com/2019/11/speed-up-pandas-4x.html
https://engineering.upside.com/a-beginners-guide-to-optimizing-pandas-code-for-speed-c09ef2c6a4d6
https://medium.com/bigdatarepublic/advanced-pandas-optimize-speed-and-memory-a654b53be6c2
https://www.thepythoncode.com/article/get-hardware-system-information-python
NOTE: I acknowledge the work of above references and this notebook uses some of code mentioned in references.