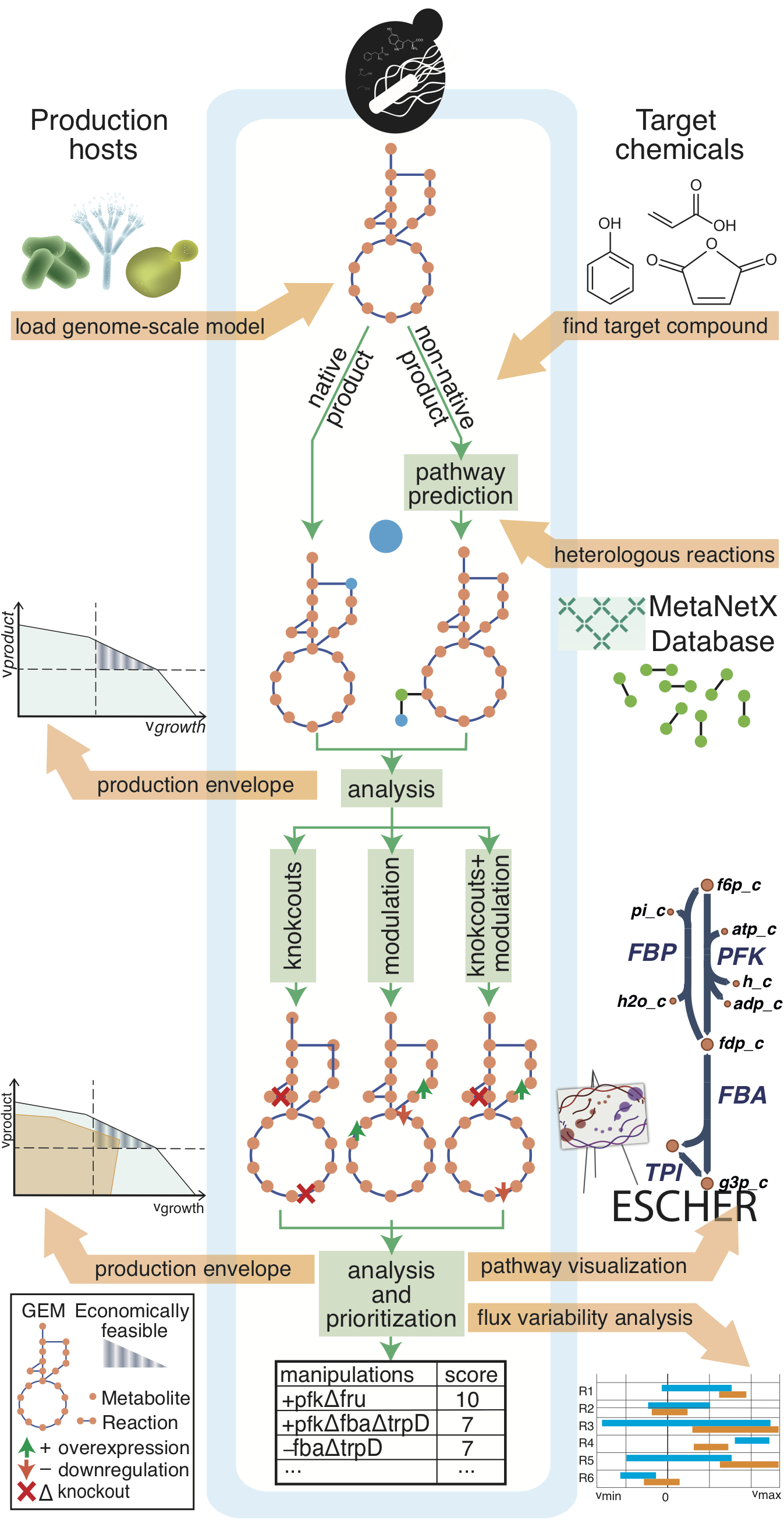
Easy strain design using a high-level interface¶
Users primarily interested in using cameo as a tool
for enumerating metabolic engineering strategies have access to cameo's advanced programming interface via cameo.api
that provides access to potential products (cameo.api.products), host organisms (cameo.api.hosts) and
a configurable design function (cameo.api.design). Running cameo.api.design requires only minimal input and will run the following workflow.
Import the advanced interface.
from cameo import api
Searching for products¶
Search by trivial name.
api.products.search('caffeine')
| InChI | SMILES | charge | formula | mass | name | source | search_rank | |
|---|---|---|---|---|---|---|---|---|
| MNXM680 | InChI=1S/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)... | CN1C=NC2=C1C(=O)N(C)C(=O)N2C | 0 | C8H10N4O2 | 194.1906 | caffeine | chebi:27732 | 0 |
Search by ChEBI ID.
api.products.search('chebi:27732')
| InChI | SMILES | charge | formula | mass | name | source | search_rank | |
|---|---|---|---|---|---|---|---|---|
| MNXM680 | InChI=1S/C8H10N4O2/c1-10-4-9-6-5(10)7(13)12(3)... | CN1C=NC2=C1C(=O)N(C)C(=O)N2C | 0 | C8H10N4O2 | 194.1906 | caffeine | chebi:27732 | 0 |
Host organisms¶
Currently the following host organisms and respective models are available in cameo. More hosts and models will be added in the future (please get in touch with us if you'd like to get a particular host organism included).
for host in api.hosts:
for model in host.models:
print(host.name, model.id)
Escherichia coli iJO1366 Saccharomyces cerevisiae iMM904
Computing strain engineering strategies¶
For demonstration purposes, we'll set a few options to limit the computational time. Also we'll create a multiprocessing view to take advantage of multicore CPUs (strain design algorithms will be run in parallel for individually predicted heterologous pathways).
from cameo.parallel import MultiprocessingView
mp_view = MultiprocessingView()
Limit the number of predicted heterlogous pathways to 4.
api.design.options.max_pathway_predictions = 4
Set a time limit of 30 minutes on individual heuristic optimizations.
api.design.options.heuristic_optimization_timeout = 30
report = api.design(product='vanillin', view=mp_view)
| Id | Name | Formula |
|---|---|---|
| MNXM754 | vanillin | C8H8O3 |
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR2795 | S-adenosyl-L-methionine + glycine <=> H(+) + S... | -1000 | 1000 |
| MNXR68718 | H2O + 3,4-dihydroxybenzoate <=> 3-dehydroshiki... | -1000 | 1000 |
Max flux: 4.29196
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR84169 | (6R)-5,10-methylenetetrahydrofolate + glycine ... | -1000 | 1000 |
| MNXR68718 | H2O + 3,4-dihydroxybenzoate <=> 3-dehydroshiki... | -1000 | 1000 |
Max flux: 7.28363
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR68718 | H2O + 3,4-dihydroxybenzoate <=> 3-dehydroshiki... | -1000 | 1000 |
| MNXR651 | 2.0 H(+) + NADH + formate <=> H2O + formaldehy... | -1000 | 1000 |
Max flux: 7.58479
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR230 | H(+) + 4-hydroxybenzoate + O2 + NADPH <=> H2O ... | -1000 | 1000 |
| MNXR640 | methanol + NAD(+) <=> H(+) + NADH + formaldehyde | -1000 | 1000 |
This is the format of your plot grid: [ (1,1) x1,y1 ] [ (1,2) x2,y2 ] [ (2,1) x3,y3 ] [ (2,2) x4,y4 ]
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR68718 | H2O + 3,4-dihydroxybenzoate <=> 3-dehydroshiki... | -1000 | 1000 |
Max flux: 3.36842
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR230 | H(+) + 4-hydroxybenzoate + O2 + NADPH <=> H2O ... | -1000 | 1000 |
Max flux: 1.90533
| equation | lower_bound | upper_bound | |
|---|---|---|---|
| MNXR4008 | H(+) + 3-oxoadipate <=> H2O + 5-oxo-4,5-dihydr... | -1000 | 1000 |
| MNXR184 | 3-oxoadipyl-CoA + succinate <=> 3-oxoadipate +... | -1000 | 1000 |
| MNXR5340 | H(+) + NADH + O2 + vanillate <=> H2O + 3,4-dih... | -1000 | 1000 |
| MNXR5336 | 2.0 H(+) + NADH + vanillate <=> H2O + vanillin... | -1000 | 1000 |
| MNXR228 | CO2 + 5-oxo-4,5-dihydro-2-furylacetate <=> H(+... | -1000 | 1000 |
| MNXR4119 | 2.0 H(+) + 3-carboxy-cis,cis-muconate <=> 3,4-... | -1000 | 1000 |
| MNXR209 | CoA + 3-oxoadipyl-CoA <=> acetyl-CoA + succiny... | -1000 | 1000 |
| MNXR3655 | 2-(carboxymethyl)-5-oxo-2,5-dihydro-2-furoate ... | -1000 | 1000 |
Max flux: 5.59223
This is the format of your plot grid: [ (1,1) x1,y1 ] [ (1,2) x2,y2 ] [ (2,1) x3,y3 ] [ (2,2) x4,y4 ]
Optimizing 6 pathways Starting optimization at Tue, 06 Feb 2018 14:06:57 Starting optimization at Tue, 06 Feb 2018 14:07:50 Starting optimization at Tue, 06 Feb 2018 14:07:57 Starting optimization at Tue, 06 Feb 2018 14:07:58
Failed to display Jupyter Widget of type HBox.
If you're reading this message in Jupyter Notebook or JupyterLab, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another notebook frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
Finished after 00:32:09 Starting optimization at Tue, 06 Feb 2018 14:39:37
Failed to display Jupyter Widget of type HBox.
If you're reading this message in Jupyter Notebook or JupyterLab, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another notebook frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
Finished after 00:33:45
Failed to display Jupyter Widget of type HBox.
If you're reading this message in Jupyter Notebook or JupyterLab, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another notebook frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
Finished after 00:33:43
Failed to display Jupyter Widget of type HBox.
If you're reading this message in Jupyter Notebook or JupyterLab, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another notebook frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
Finished after 00:33:43 Starting optimization at Tue, 06 Feb 2018 14:41:56
Failed to display Jupyter Widget of type HBox.
If you're reading this message in Jupyter Notebook or JupyterLab, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another notebook frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
Failed to display Jupyter Widget of type HBox.
If you're reading this message in Jupyter Notebook or JupyterLab, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another notebook frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
Finished after 00:30:09 Finished after 00:30:02
/Users/joaca/Documents/repositories/cameo/cameo/strain_design/heuristic/evolutionary/objective_functions.py:287: RuntimeWarning: invalid value encountered in double_scalars /Users/joaca/Documents/repositories/cameo/cameo/strain_design/heuristic/evolutionary/objective_functions.py:354: RuntimeWarning: invalid value encountered in double_scalars
report
| host | model | manipulations | heterologous_pathway | fitness | yield | product | biomass | method | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | Saccharomyces cerevisiae | iMM904 | (-YPL110C) | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.000043 | 0.000199 | 0.001489 | 2.879114e-01 | PathwayPredictor+OptGene |
| 1 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.572262 | 4.291964 | 8.136452e-08 | PathwayPredictor+DifferentialFVA |
| 2 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.000000 | 0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 3 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.01699), reaction.3OA... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.508185 | 3.351657 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 4 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.025534), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.093738 | 0.381679 | 2.862596 | 3.274580e-01 | PathwayPredictor+DifferentialFVA |
| 5 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.034045), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.475180 | 3.563847 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 6 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.042474), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.104223 | 0.254625 | 1.909686 | 5.457622e-01 | PathwayPredictor+DifferentialFVA |
| 7 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.050969), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.093864 | 0.191097 | 1.433230 | 6.549146e-01 | PathwayPredictor+DifferentialFVA |
| 8 | Escherichia coli | iJO1366 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.000000 | 0.000000 | 9.823718e-01 | PathwayPredictor+DifferentialFVA |
| 9 | Escherichia coli | iJO1366 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | -0.000000 | 0.000000 | 0.000000 | 9.823718e-01 | PathwayPredictor+DifferentialFVA |
| 10 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.971150 | 7.283628 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 11 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | 0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 12 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | 0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 13 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | -0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 14 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | 0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 15 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.042474), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.178214 | 0.435389 | 3.265417 | 5.457622e-01 | PathwayPredictor+DifferentialFVA |
| 16 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.050969), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.160501 | 0.326762 | 2.450714 | 6.549146e-01 | PathwayPredictor+DifferentialFVA |
| 17 | Escherichia coli | iJO1366 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.000000 | 0.000000 | 9.823718e-01 | PathwayPredictor+DifferentialFVA |
| 18 | Escherichia coli | iJO1366 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | -0.000000 | 0.000000 | 0.000000 | 9.823718e-01 | PathwayPredictor+DifferentialFVA |
| 19 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 1.011305 | 7.584790 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 20 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | -0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 21 | Escherichia coli | iJO1366 | (-reaction.3OAR140, -reaction.3OAS140, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | 0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 22 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.025485), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 23 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.034045), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.668001 | 4.390359 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 24 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.042474), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.185581 | 0.453387 | 3.400405 | 5.457622e-01 | PathwayPredictor+DifferentialFVA |
| 25 | Escherichia coli | iJO1366 | (reaction.3OAR140(value=0.050969), reaction.3O... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.167136 | 0.340270 | 2.552028 | 6.549146e-01 | PathwayPredictor+DifferentialFVA |
| 26 | Escherichia coli | iJO1366 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | -0.000000 | 0.000000 | 0.000000 | 9.823718e-01 | PathwayPredictor+DifferentialFVA |
| 27 | Escherichia coli | iJO1366 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | -0.000000 | 0.000000 | 0.000000 | 9.823718e-01 | PathwayPredictor+DifferentialFVA |
| 28 | Saccharomyces cerevisiae | iMM904 | (-reaction.13GS, -reaction.2OXOADPtim, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.449123 | 3.368421 | 5.205722e-09 | PathwayPredictor+DifferentialFVA |
| 29 | Saccharomyces cerevisiae | iMM904 | (-reaction.13GS, -reaction.2OXOADPtim, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | 0.000000 | 4.984455e-12 | PathwayPredictor+DifferentialFVA |
| 30 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.072594), reaction.2DDA7... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 31 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.108891), reaction.2DDA7... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 32 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.145187), reaction.2DDA7... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 33 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.181484), reaction.2DDA7... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 34 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.217781), reaction.2DDA7... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 35 | Saccharomyces cerevisiae | iMM904 | (-reaction.SULR, -reaction.THRS, -reaction.TMD... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | -0.000000 | -0.000000 | 0.000000e+00 | PathwayPredictor+DifferentialFVA |
| 36 | Saccharomyces cerevisiae | iMM904 | (-reaction.ENO, -reaction.ERGSTter, -reaction.... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 37 | Saccharomyces cerevisiae | iMM904 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | -0.000000 | 0.000000 | 0.000000 | 2.878657e-01 | PathwayPredictor+DifferentialFVA |
| 38 | Saccharomyces cerevisiae | iMM904 | (-reaction.13GS, -reaction.2OXOADPtim, -reacti... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.000000 | 0.254043 | 1.905325 | 4.603305e-08 | PathwayPredictor+DifferentialFVA |
| 39 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.036297), reaction.2OXOA... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.005438 | 0.226674 | 1.700055 | 3.198537e-02 | PathwayPredictor+DifferentialFVA |
| 40 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.072594), reaction.2OXOA... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.009562 | 0.199305 | 1.494785 | 6.397013e-02 | PathwayPredictor+DifferentialFVA |
| 41 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.108891), reaction.2OXOA... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.012374 | 0.171935 | 1.289514 | 9.595523e-02 | PathwayPredictor+DifferentialFVA |
| 42 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.145187), reaction.2OXOA... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.013872 | 0.144566 | 1.084244 | 1.279406e-01 | PathwayPredictor+DifferentialFVA |
| 43 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.181484), reaction.2OXOA... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.014057 | 0.117197 | 0.878974 | 1.599255e-01 | PathwayPredictor+DifferentialFVA |
| 44 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.21778), reaction.2OXOAD... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | 0.012757 | 0.088634 | 0.664755 | 1.919105e-01 | PathwayPredictor+DifferentialFVA |
| 45 | Saccharomyces cerevisiae | iMM904 | () | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | -0.000000 | -0.000000 | -0.000000 | 2.878657e-01 | PathwayPredictor+DifferentialFVA |
| 46 | Saccharomyces cerevisiae | iMM904 | (-reaction.ACOTAim, -reaction.ACS, -reaction.A... | (+reaction.MNXR5340#metanetx:MNXR5340, +reacti... | NaN | NaN | -0.000000 | -1.316513e-12 | PathwayPredictor+DifferentialFVA |
| 47 | Saccharomyces cerevisiae | iMM904 | (-reaction.13GS, -reaction.2OXOADPtim, -reacti... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | -0.000000 | 0.745631 | 5.592233 | -7.750803e-11 | PathwayPredictor+DifferentialFVA |
| 48 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.036303), reaction.2OXOA... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.015942 | 0.664458 | 4.983433 | 3.199022e-02 | PathwayPredictor+DifferentialFVA |
| 49 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.072605), reaction.2OXOA... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.027989 | 0.583284 | 4.374633 | 6.398044e-02 | PathwayPredictor+DifferentialFVA |
| 50 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.108908), reaction.2OXOA... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.036141 | 0.502111 | 3.765833 | 9.597049e-02 | PathwayPredictor+DifferentialFVA |
| 51 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.14521), reaction.2OXOAD... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.040398 | 0.420938 | 3.157035 | 1.279606e-01 | PathwayPredictor+DifferentialFVA |
| 52 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.181513), reaction.2OXOA... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.040759 | 0.339765 | 2.548235 | 1.599507e-01 | PathwayPredictor+DifferentialFVA |
| 53 | Saccharomyces cerevisiae | iMM904 | (reaction.13GS(value=0.217815), reaction.2OXOA... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.037226 | 0.258591 | 1.939434 | 1.919409e-01 | PathwayPredictor+DifferentialFVA |
| 54 | Saccharomyces cerevisiae | iMM904 | () | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.000043 | 0.000199 | 0.001489 | 2.879114e-01 | PathwayPredictor+DifferentialFVA |
| 55 | Saccharomyces cerevisiae | iMM904 | (-reaction.GLCt1, -reaction.GLNS, -reaction.GL... | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | NaN | NaN | NaN | NaN | PathwayPredictor+DifferentialFVA |
| 56 | Saccharomyces cerevisiae | iMM904 | () | (+reaction.MNXR4008#metanetx:MNXR4008, +reacti... | 0.000043 | 0.000199 | 0.001489 | 2.879114e-01 | PathwayPredictor+DifferentialFVA |
IPython notebook¶
Click here to download this page as an IPython notebook.