Intro to Random Forests¶
About this course¶
Teaching approach¶
This course is being taught by Jeremy Howard, and was developed by Jeremy along with Rachel Thomas. Rachel has been dealing with a life-threatening illness so will not be teaching as originally planned this year.
Jeremy has worked in a number of different areas - feel free to ask about anything that he might be able to help you with at any time, even if not directly related to the current topic:
- Management consultant (McKinsey; AT Kearney)
- Self-funded startup entrepreneur (Fastmail: first consumer synchronized email; Optimal Decisions: first optimized insurance pricing)
- VC-funded startup entrepreneur: (Kaggle; Enlitic: first deep-learning medical company)
I'll be using a top-down teaching method, which is different from how most math courses operate. Typically, in a bottom-up approach, you first learn all the separate components you will be using, and then you gradually build them up into more complex structures. The problems with this are that students often lose motivation, don't have a sense of the "big picture", and don't know what they'll need.
If you took the fast.ai deep learning course, that is what we used. You can hear more about my teaching philosophy in this blog post or in this talk.
Harvard Professor David Perkins has a book, Making Learning Whole in which he uses baseball as an analogy. We don't require kids to memorize all the rules of baseball and understand all the technical details before we let them play the game. Rather, they start playing with a just general sense of it, and then gradually learn more rules/details as time goes on.
All that to say, don't worry if you don't understand everything at first! You're not supposed to. We will start using some "black boxes" such as random forests that haven't yet been explained in detail, and then we'll dig into the lower level details later.
To start, focus on what things DO, not what they ARE.
Your practice¶
People learn by:
- doing (coding and building)
- explaining what they've learned (by writing or helping others)
Therefore, we suggest that you practice these skills on Kaggle by:
- Entering competitions (doing)
- Creating Kaggle kernels (explaining)
It's OK if you don't get good competition ranks or any kernel votes at first - that's totally normal! Just try to keep improving every day, and you'll see the results over time.
To get better at technical writing, study the top ranked Kaggle kernels from past competitions, and read posts from well-regarded technical bloggers. Some good role models include:
- Peter Norvig (more here)
- Stephen Merity
- Julia Evans (more here)
- Julia Ferraioli
- Edwin Chen
- Slav Ivanov (fast.ai student)
- Brad Kenstler (fast.ai and USF MSAN student)
Books¶
The more familiarity you have with numeric programming in Python, the better. If you're looking to improve in this area, we strongly suggest Wes McKinney's Python for Data Analysis, 2nd ed.
For machine learning with Python, we recommend:
- Introduction to Machine Learning with Python: From one of the scikit-learn authors, which is the main library we'll be using
- Python Machine Learning: Machine Learning and Deep Learning with Python, scikit-learn, and TensorFlow, 2nd Edition: New version of a very successful book. A lot of the new material however covers deep learning in Tensorflow, which isn't relevant to this course
- Hands-On Machine Learning with Scikit-Learn and TensorFlow
Syllabus in brief¶
Depending on time and class interests, we'll cover something like (not necessarily in this order):
- Train vs test
- Effective validation set construction
- Trees and ensembles
- Creating random forests
- Interpreting random forests
- What is ML? Why do we use it?
- What makes a good ML project?
- Structured vs unstructured data
- Examples of failures/mistakes
- Feature engineering
- Domain specific - dates, URLs, text
- Embeddings / latent factors
- Regularized models trained with SGD
- GLMs, Elasticnet, etc (NB: see what James covered)
- Basic neural nets
- PyTorch
- Broadcasting, Matrix Multiplication
- Training loop, backpropagation
- KNN
- CV / bootstrap (Diabetes data set?)
- Ethical considerations
Skip:
- Dimensionality reduction
- Interactions
- Monitoring training
- Collaborative filtering
- Momentum and LR annealing
Imports¶
%load_ext autoreload
%autoreload 2
%matplotlib inline
from fastai.imports import *
from fastai.structured import *
from pandas_summary import DataFrameSummary
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from IPython.display import display
from sklearn import metrics
/home/jhoward/anaconda3/lib/python3.6/site-packages/sklearn/ensemble/weight_boosting.py:29: DeprecationWarning: numpy.core.umath_tests is an internal NumPy module and should not be imported. It will be removed in a future NumPy release. from numpy.core.umath_tests import inner1d
PATH = "data/bulldozers/"
!ls {PATH}
Data%20Dictionary.xlsx Test.csv Train.csv Machine_Appendix.csv tmp Valid.csv median_benchmark.csv TrainAndValid.7z ValidSolution.csv models TrainAndValid.csv random_forest_benchmark_test.csv TrainAndValid.zip
Introduction to Blue Book for Bulldozers¶
About...¶
...our teaching¶
At fast.ai we have a distinctive teaching philosophy of "the whole game". This is different from how most traditional math & technical courses are taught, where you have to learn all the individual elements before you can combine them (Harvard professor David Perkins call this elementitis), but it is similar to how topics like driving and baseball are taught. That is, you can start driving without knowing how an internal combustion engine works, and children begin playing baseball before they learn all the formal rules.
...our approach to machine learning¶
Most machine learning courses will throw at you dozens of different algorithms, with a brief technical description of the math behind them, and maybe a toy example. You're left confused by the enormous range of techniques shown and have little practical understanding of how to apply them.
The good news is that modern machine learning can be distilled down to a couple of key techniques that are of very wide applicability. Recent studies have shown that the vast majority of datasets can be best modeled with just two methods:
- Ensembles of decision trees (i.e. Random Forests and Gradient Boosting Machines), mainly for structured data (such as you might find in a database table at most companies)
- Multi-layered neural networks learnt with SGD (i.e. shallow and/or deep learning), mainly for unstructured data (such as audio, vision, and natural language)
In this course we'll be doing a deep dive into random forests, and simple models learnt with SGD. You'll be learning about gradient boosting and deep learning in part 2.
...this dataset¶
We will be looking at the Blue Book for Bulldozers Kaggle Competition: "The goal of the contest is to predict the sale price of a particular piece of heavy equiment at auction based on it's usage, equipment type, and configuration. The data is sourced from auction result postings and includes information on usage and equipment configurations."
This is a very common type of dataset and prediciton problem, and similar to what you may see in your project or workplace.
...Kaggle Competitions¶
Kaggle is an awesome resource for aspiring data scientists or anyone looking to improve their machine learning skills. There is nothing like being able to get hands-on practice and receiving real-time feedback to help you improve your skills.
Kaggle provides:
- Interesting data sets
- Feedback on how you're doing
- A leader board to see what's good, what's possible, and what's state-of-art.
- Blog posts by winning contestants share useful tips and techniques.
The data¶
Look at the data¶
Kaggle provides info about some of the fields of our dataset; on the Kaggle Data info page they say the following:
For this competition, you are predicting the sale price of bulldozers sold at auctions. The data for this competition is split into three parts:
- Train.csv is the training set, which contains data through the end of 2011.
- Valid.csv is the validation set, which contains data from January 1, 2012 - April 30, 2012. You make predictions on this set throughout the majority of the competition. Your score on this set is used to create the public leaderboard.
- Test.csv is the test set, which won't be released until the last week of the competition. It contains data from May 1, 2012 - November 2012. Your score on the test set determines your final rank for the competition.
The key fields are in train.csv are:
- SalesID: the unique identifier of the sale
- MachineID: the unique identifier of a machine. A machine can be sold multiple times
- saleprice: what the machine sold for at auction (only provided in train.csv)
- saledate: the date of the sale
Question
What stands out to you from the above description? What needs to be true of our training and validation sets?
df_raw = pd.read_csv(f'{PATH}Train.csv', low_memory=False,
parse_dates=["saledate"])
In any sort of data science work, it's important to look at your data, to make sure you understand the format, how it's stored, what type of values it holds, etc. Even if you've read descriptions about your data, the actual data may not be what you expect.
def display_all(df):
with pd.option_context("display.max_rows", 1000, "display.max_columns", 1000):
display(df)
display_all(df_raw.tail().T)
| 401120 | 401121 | 401122 | 401123 | 401124 | |
|---|---|---|---|---|---|
| SalesID | 6333336 | 6333337 | 6333338 | 6333341 | 6333342 |
| SalePrice | 10500 | 11000 | 11500 | 9000 | 7750 |
| MachineID | 1840702 | 1830472 | 1887659 | 1903570 | 1926965 |
| ModelID | 21439 | 21439 | 21439 | 21435 | 21435 |
| datasource | 149 | 149 | 149 | 149 | 149 |
| auctioneerID | 1 | 1 | 1 | 2 | 2 |
| YearMade | 2005 | 2005 | 2005 | 2005 | 2005 |
| MachineHoursCurrentMeter | NaN | NaN | NaN | NaN | NaN |
| UsageBand | NaN | NaN | NaN | NaN | NaN |
| saledate | 2011-11-02 00:00:00 | 2011-11-02 00:00:00 | 2011-11-02 00:00:00 | 2011-10-25 00:00:00 | 2011-10-25 00:00:00 |
| fiModelDesc | 35NX2 | 35NX2 | 35NX2 | 30NX | 30NX |
| fiBaseModel | 35 | 35 | 35 | 30 | 30 |
| fiSecondaryDesc | NX | NX | NX | NX | NX |
| fiModelSeries | 2 | 2 | 2 | NaN | NaN |
| fiModelDescriptor | NaN | NaN | NaN | NaN | NaN |
| ProductSize | Mini | Mini | Mini | Mini | Mini |
| fiProductClassDesc | Hydraulic Excavator, Track - 3.0 to 4.0 Metric... | Hydraulic Excavator, Track - 3.0 to 4.0 Metric... | Hydraulic Excavator, Track - 3.0 to 4.0 Metric... | Hydraulic Excavator, Track - 2.0 to 3.0 Metric... | Hydraulic Excavator, Track - 2.0 to 3.0 Metric... |
| state | Maryland | Maryland | Maryland | Florida | Florida |
| ProductGroup | TEX | TEX | TEX | TEX | TEX |
| ProductGroupDesc | Track Excavators | Track Excavators | Track Excavators | Track Excavators | Track Excavators |
| Drive_System | NaN | NaN | NaN | NaN | NaN |
| Enclosure | EROPS | EROPS | EROPS | EROPS | EROPS |
| Forks | NaN | NaN | NaN | NaN | NaN |
| Pad_Type | NaN | NaN | NaN | NaN | NaN |
| Ride_Control | NaN | NaN | NaN | NaN | NaN |
| Stick | NaN | NaN | NaN | NaN | NaN |
| Transmission | NaN | NaN | NaN | NaN | NaN |
| Turbocharged | NaN | NaN | NaN | NaN | NaN |
| Blade_Extension | NaN | NaN | NaN | NaN | NaN |
| Blade_Width | NaN | NaN | NaN | NaN | NaN |
| Enclosure_Type | NaN | NaN | NaN | NaN | NaN |
| Engine_Horsepower | NaN | NaN | NaN | NaN | NaN |
| Hydraulics | Auxiliary | Standard | Auxiliary | Standard | Standard |
| Pushblock | NaN | NaN | NaN | NaN | NaN |
| Ripper | NaN | NaN | NaN | NaN | NaN |
| Scarifier | NaN | NaN | NaN | NaN | NaN |
| Tip_Control | NaN | NaN | NaN | NaN | NaN |
| Tire_Size | NaN | NaN | NaN | NaN | NaN |
| Coupler | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified |
| Coupler_System | NaN | NaN | NaN | NaN | NaN |
| Grouser_Tracks | NaN | NaN | NaN | NaN | NaN |
| Hydraulics_Flow | NaN | NaN | NaN | NaN | NaN |
| Track_Type | Steel | Steel | Steel | Steel | Steel |
| Undercarriage_Pad_Width | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified |
| Stick_Length | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified |
| Thumb | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified |
| Pattern_Changer | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified | None or Unspecified |
| Grouser_Type | Double | Double | Double | Double | Double |
| Backhoe_Mounting | NaN | NaN | NaN | NaN | NaN |
| Blade_Type | NaN | NaN | NaN | NaN | NaN |
| Travel_Controls | NaN | NaN | NaN | NaN | NaN |
| Differential_Type | NaN | NaN | NaN | NaN | NaN |
| Steering_Controls | NaN | NaN | NaN | NaN | NaN |
display_all(df_raw.describe(include='all').T)
| count | unique | top | freq | first | last | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SalesID | 401125 | NaN | NaN | NaN | NaN | NaN | 1.91971e+06 | 909021 | 1.13925e+06 | 1.41837e+06 | 1.63942e+06 | 2.24271e+06 | 6.33334e+06 |
| SalePrice | 401125 | NaN | NaN | NaN | NaN | NaN | 31099.7 | 23036.9 | 4750 | 14500 | 24000 | 40000 | 142000 |
| MachineID | 401125 | NaN | NaN | NaN | NaN | NaN | 1.2179e+06 | 440992 | 0 | 1.0887e+06 | 1.27949e+06 | 1.46807e+06 | 2.48633e+06 |
| ModelID | 401125 | NaN | NaN | NaN | NaN | NaN | 6889.7 | 6221.78 | 28 | 3259 | 4604 | 8724 | 37198 |
| datasource | 401125 | NaN | NaN | NaN | NaN | NaN | 134.666 | 8.96224 | 121 | 132 | 132 | 136 | 172 |
| auctioneerID | 380989 | NaN | NaN | NaN | NaN | NaN | 6.55604 | 16.9768 | 0 | 1 | 2 | 4 | 99 |
| YearMade | 401125 | NaN | NaN | NaN | NaN | NaN | 1899.16 | 291.797 | 1000 | 1985 | 1995 | 2000 | 2013 |
| MachineHoursCurrentMeter | 142765 | NaN | NaN | NaN | NaN | NaN | 3457.96 | 27590.3 | 0 | 0 | 0 | 3025 | 2.4833e+06 |
| UsageBand | 69639 | 3 | Medium | 33985 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| saledate | 401125 | 3919 | 2009-02-16 00:00:00 | 1932 | 1989-01-17 00:00:00 | 2011-12-30 00:00:00 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fiModelDesc | 401125 | 4999 | 310G | 5039 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fiBaseModel | 401125 | 1950 | 580 | 19798 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fiSecondaryDesc | 263934 | 175 | C | 43235 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fiModelSeries | 56908 | 122 | II | 13202 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fiModelDescriptor | 71919 | 139 | L | 15875 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ProductSize | 190350 | 6 | Medium | 62274 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| fiProductClassDesc | 401125 | 74 | Backhoe Loader - 14.0 to 15.0 Ft Standard Digg... | 56166 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| state | 401125 | 53 | Florida | 63944 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ProductGroup | 401125 | 6 | TEX | 101167 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ProductGroupDesc | 401125 | 6 | Track Excavators | 101167 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Drive_System | 104361 | 4 | Two Wheel Drive | 46139 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Enclosure | 400800 | 6 | OROPS | 173932 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Forks | 192077 | 2 | None or Unspecified | 178300 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Pad_Type | 79134 | 4 | None or Unspecified | 70614 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Ride_Control | 148606 | 3 | No | 77685 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Stick | 79134 | 2 | Standard | 48829 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Transmission | 183230 | 8 | Standard | 140328 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Turbocharged | 79134 | 2 | None or Unspecified | 75211 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Blade_Extension | 25219 | 2 | None or Unspecified | 24692 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Blade_Width | 25219 | 6 | 14' | 9615 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Enclosure_Type | 25219 | 3 | None or Unspecified | 21923 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Engine_Horsepower | 25219 | 2 | No | 23937 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Hydraulics | 320570 | 12 | 2 Valve | 141404 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Pushblock | 25219 | 2 | None or Unspecified | 19463 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Ripper | 104137 | 4 | None or Unspecified | 83452 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Scarifier | 25230 | 2 | None or Unspecified | 12719 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Tip_Control | 25219 | 3 | None or Unspecified | 16207 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Tire_Size | 94718 | 17 | None or Unspecified | 46339 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Coupler | 213952 | 3 | None or Unspecified | 184582 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Coupler_System | 43458 | 2 | None or Unspecified | 40430 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Grouser_Tracks | 43362 | 2 | None or Unspecified | 40515 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Hydraulics_Flow | 43362 | 3 | Standard | 42784 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Track_Type | 99153 | 2 | Steel | 84880 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Undercarriage_Pad_Width | 99872 | 19 | None or Unspecified | 79651 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Stick_Length | 99218 | 29 | None or Unspecified | 78820 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Thumb | 99288 | 3 | None or Unspecified | 83093 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Pattern_Changer | 99218 | 3 | None or Unspecified | 90255 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Grouser_Type | 99153 | 3 | Double | 84653 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Backhoe_Mounting | 78672 | 2 | None or Unspecified | 78652 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Blade_Type | 79833 | 10 | PAT | 38612 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Travel_Controls | 79834 | 7 | None or Unspecified | 69923 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Differential_Type | 69411 | 4 | Standard | 68073 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| Steering_Controls | 69369 | 5 | Conventional | 68679 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
It's important to note what metric is being used for a project. Generally, selecting the metric(s) is an important part of the project setup. However, in this case Kaggle tells us what metric to use: RMSLE (root mean squared log error) between the actual and predicted auction prices. Therefore we take the log of the prices, so that RMSE will give us what we need.
df_raw.SalePrice = np.log(df_raw.SalePrice)
Initial processing¶
m = RandomForestRegressor(n_jobs=-1)
# The following code is supposed to fail due to string values in the input data
m.fit(df_raw.drop('SalePrice', axis=1), df_raw.SalePrice)
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) <ipython-input-10-6e70335c9573> in <module>() 1 m = RandomForestRegressor(n_jobs=-1) 2 # The following code is supposed to fail due to string values in the input data ----> 3 m.fit(df_raw.drop('SalePrice', axis=1), df_raw.SalePrice) ~/anaconda3/lib/python3.6/site-packages/sklearn/ensemble/forest.py in fit(self, X, y, sample_weight) 245 """ 246 # Validate or convert input data --> 247 X = check_array(X, accept_sparse="csc", dtype=DTYPE) 248 y = check_array(y, accept_sparse='csc', ensure_2d=False, dtype=None) 249 if sample_weight is not None: ~/anaconda3/lib/python3.6/site-packages/sklearn/utils/validation.py in check_array(array, accept_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, ensure_min_samples, ensure_min_features, warn_on_dtype, estimator) 431 force_all_finite) 432 else: --> 433 array = np.array(array, dtype=dtype, order=order, copy=copy) 434 435 if ensure_2d: ValueError: could not convert string to float: 'Conventional'
This dataset contains a mix of continuous and categorical variables.
The following method extracts particular date fields from a complete datetime for the purpose of constructing categoricals. You should always consider this feature extraction step when working with date-time. Without expanding your date-time into these additional fields, you can't capture any trend/cyclical behavior as a function of time at any of these granularities.
add_datepart(df_raw, 'saledate')
df_raw.saleYear.head()
0 2006 1 2004 2 2004 3 2011 4 2009 Name: saleYear, dtype: int64
The categorical variables are currently stored as strings, which is inefficient, and doesn't provide the numeric coding required for a random forest. Therefore we call train_cats to convert strings to pandas categories.
train_cats(df_raw)
We can specify the order to use for categorical variables if we wish:
df_raw.UsageBand.cat.categories
Index(['High', 'Low', 'Medium'], dtype='object')
df_raw.UsageBand.cat.set_categories(['High', 'Medium', 'Low'], ordered=True, inplace=True)
Normally, pandas will continue displaying the text categories, while treating them as numerical data internally. Optionally, we can replace the text categories with numbers, which will make this variable non-categorical, like so:.
df_raw.UsageBand = df_raw.UsageBand.cat.codes
We're still not quite done - for instance we have lots of missing values, which we can't pass directly to a random forest.
display_all(df_raw.isnull().sum().sort_index()/len(df_raw))
Backhoe_Mounting 0.803872 Blade_Extension 0.937129 Blade_Type 0.800977 Blade_Width 0.937129 Coupler 0.466620 Coupler_System 0.891660 Differential_Type 0.826959 Drive_System 0.739829 Enclosure 0.000810 Enclosure_Type 0.937129 Engine_Horsepower 0.937129 Forks 0.521154 Grouser_Tracks 0.891899 Grouser_Type 0.752813 Hydraulics 0.200823 Hydraulics_Flow 0.891899 MachineHoursCurrentMeter 0.644089 MachineID 0.000000 ModelID 0.000000 Pad_Type 0.802720 Pattern_Changer 0.752651 ProductGroup 0.000000 ProductGroupDesc 0.000000 ProductSize 0.525460 Pushblock 0.937129 Ride_Control 0.629527 Ripper 0.740388 SalePrice 0.000000 SalesID 0.000000 Scarifier 0.937102 Steering_Controls 0.827064 Stick 0.802720 Stick_Length 0.752651 Thumb 0.752476 Tip_Control 0.937129 Tire_Size 0.763869 Track_Type 0.752813 Transmission 0.543210 Travel_Controls 0.800975 Turbocharged 0.802720 Undercarriage_Pad_Width 0.751020 UsageBand 0.000000 YearMade 0.000000 auctioneerID 0.050199 datasource 0.000000 fiBaseModel 0.000000 fiModelDesc 0.000000 fiModelDescriptor 0.820707 fiModelSeries 0.858129 fiProductClassDesc 0.000000 fiSecondaryDesc 0.342016 saleDay 0.000000 saleDayofweek 0.000000 saleDayofyear 0.000000 saleElapsed 0.000000 saleIs_month_end 0.000000 saleIs_month_start 0.000000 saleIs_quarter_end 0.000000 saleIs_quarter_start 0.000000 saleIs_year_end 0.000000 saleIs_year_start 0.000000 saleMonth 0.000000 saleWeek 0.000000 saleYear 0.000000 state 0.000000 dtype: float64
But let's save this file for now, since it's already in format can we be stored and accessed efficiently.
os.makedirs('tmp', exist_ok=True)
df_raw.to_feather('tmp/bulldozers-raw')
Pre-processing¶
In the future we can simply read it from this fast format.
df_raw = pd.read_feather('tmp/bulldozers-raw')
We'll replace categories with their numeric codes, handle missing continuous values, and split the dependent variable into a separate variable.
df, y, nas = proc_df(df_raw, 'SalePrice')
We now have something we can pass to a random forest!
m = RandomForestRegressor(n_jobs=-1)
m.fit(df, y)
m.score(df,y)
0.9830962179413453
In statistics, the coefficient of determination, denoted R2 or r2 and pronounced "R squared", is the proportion of the variance in the dependent variable that is predictable from the independent variable(s). https://en.wikipedia.org/wiki/Coefficient_of_determination
Wow, an r^2 of 0.98 - that's great, right? Well, perhaps not...
Possibly the most important idea in machine learning is that of having separate training & validation data sets. As motivation, suppose you don't divide up your data, but instead use all of it. And suppose you have lots of parameters:

The error for the pictured data points is lowest for the model on the far right (the blue curve passes through the red points almost perfectly), yet it's not the best choice. Why is that? If you were to gather some new data points, they most likely would not be on that curve in the graph on the right, but would be closer to the curve in the middle graph.
This illustrates how using all our data can lead to overfitting. A validation set helps diagnose this problem.
def split_vals(a,n): return a[:n].copy(), a[n:].copy()
n_valid = 12000 # same as Kaggle's test set size
n_trn = len(df)-n_valid
raw_train, raw_valid = split_vals(df_raw, n_trn)
X_train, X_valid = split_vals(df, n_trn)
y_train, y_valid = split_vals(y, n_trn)
X_train.shape, y_train.shape, X_valid.shape
((389125, 66), (389125,), (12000, 66))
Random Forests¶
Base model¶
Let's try our model again, this time with separate training and validation sets.
def rmse(x,y): return math.sqrt(((x-y)**2).mean())
def print_score(m):
res = [rmse(m.predict(X_train), y_train), rmse(m.predict(X_valid), y_valid),
m.score(X_train, y_train), m.score(X_valid, y_valid)]
if hasattr(m, 'oob_score_'): res.append(m.oob_score_)
print(res)
m = RandomForestRegressor(n_jobs=-1)
%time m.fit(X_train, y_train)
print_score(m)
CPU times: user 1min 4s, sys: 372 ms, total: 1min 4s Wall time: 8.56 s [0.0904611534175684, 0.2517003033389636, 0.9828975209204237, 0.8868601882297901]
An r^2 in the high-80's isn't bad at all (and the RMSLE puts us around rank 100 of 470 on the Kaggle leaderboard), but we can see from the validation set score that we're over-fitting badly. To understand this issue, let's simplify things down to a single small tree.
Speeding things up¶
df_trn, y_trn, nas = proc_df(df_raw, 'SalePrice', subset=30000, na_dict=nas)
X_train, _ = split_vals(df_trn, 20000)
y_train, _ = split_vals(y_trn, 20000)
m = RandomForestRegressor(n_jobs=-1)
%time m.fit(X_train, y_train)
print_score(m)
CPU times: user 6.45 s, sys: 132 ms, total: 6.58 s Wall time: 539 ms [0.11129876897034846, 0.3501178589366683, 0.9730338701182212, 0.7810845051996299]
Single tree¶
m = RandomForestRegressor(n_estimators=1, max_depth=3, bootstrap=False, n_jobs=-1)
m.fit(X_train, y_train)
print_score(m)
[0.4965829795739235, 0.5246832258551836, 0.50149617735615859, 0.5083655198087873]
draw_tree(m.estimators_[0], df_trn, precision=3)
Let's see what happens if we create a bigger tree.
m = RandomForestRegressor(n_estimators=1, bootstrap=False, n_jobs=-1)
m.fit(X_train, y_train)
print_score(m)
[6.526751786450488e-17, 0.38473652894699306, 1.0, 0.73565273648797624]
The training set result looks great! But the validation set is worse than our original model. This is why we need to use bagging of multiple trees to get more generalizable results.
Bagging¶
Intro to bagging¶
To learn about bagging in random forests, let's start with our basic model again.
m = RandomForestRegressor(n_jobs=-1)
m.fit(X_train, y_train)
print_score(m)
[0.11745691160954547, 0.27959279688230376, 0.97139456205050101, 0.86039533492251219]
We'll grab the predictions for each individual tree, and look at one example.
preds = np.stack([t.predict(X_valid) for t in m.estimators_])
preds[:,0], np.mean(preds[:,0]), y_valid[0]
(array([ 9.21034, 8.9872 , 8.9872 , 8.9872 , 8.9872 , 9.21034, 8.92266, 9.21034, 9.21034, 8.9872 ]), 9.0700003890739005, 9.1049798563183568)
preds.shape
(10, 12000)
plt.plot([metrics.r2_score(y_valid, np.mean(preds[:i+1], axis=0)) for i in range(10)]);
The shape of this curve suggests that adding more trees isn't going to help us much. Let's check. (Compare this to our original model on a sample)
m = RandomForestRegressor(n_estimators=20, n_jobs=-1)
m.fit(X_train, y_train)
print_score(m)
[0.10721195540628872, 0.2777358026154778, 0.9761670456844791, 0.86224362387001874]
m = RandomForestRegressor(n_estimators=40, n_jobs=-1)
m.fit(X_train, y_train)
print_score(m)
[0.1029319603663909, 0.2725488716109634, 0.97803192843821529, 0.86734099039701873]
m = RandomForestRegressor(n_estimators=80, n_jobs=-1)
m.fit(X_train, y_train)
print_score(m)
[0.09942284423261978, 0.27026457977935875, 0.97950425012208453, 0.86955536025947799]
Out-of-bag (OOB) score¶
Is our validation set worse than our training set because we're over-fitting, or because the validation set is for a different time period, or a bit of both? With the existing information we've shown, we can't tell. However, random forests have a very clever trick called out-of-bag (OOB) error which can handle this (and more!)
The idea is to calculate error on the training set, but only include the trees in the calculation of a row's error where that row was not included in training that tree. This allows us to see whether the model is over-fitting, without needing a separate validation set.
This also has the benefit of allowing us to see whether our model generalizes, even if we only have a small amount of data so want to avoid separating some out to create a validation set.
This is as simple as adding one more parameter to our model constructor. We print the OOB error last in our print_score function below.
m = RandomForestRegressor(n_estimators=40, n_jobs=-1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
[0.10198464613020647, 0.2714485881623037, 0.9786192457999483, 0.86840992079038759, 0.84831537630038534]
This shows that our validation set time difference is making an impact, as is model over-fitting.
Reducing over-fitting¶
Subsampling¶
It turns out that one of the easiest ways to avoid over-fitting is also one of the best ways to speed up analysis: subsampling. Let's return to using our full dataset, so that we can demonstrate the impact of this technique.
df_trn, y_trn, nas = proc_df(df_raw, 'SalePrice')
X_train, X_valid = split_vals(df_trn, n_trn)
y_train, y_valid = split_vals(y_trn, n_trn)
The basic idea is this: rather than limit the total amount of data that our model can access, let's instead limit it to a different random subset per tree. That way, given enough trees, the model can still see all the data, but for each individual tree it'll be just as fast as if we had cut down our dataset as before.
set_rf_samples(20000)
m = RandomForestRegressor(n_jobs=-1, oob_score=True)
%time m.fit(X_train, y_train)
print_score(m)
CPU times: user 8.38 s, sys: 428 ms, total: 8.81 s Wall time: 3.49 s [0.24021020451254516, 0.2780622994610262, 0.87937208432405256, 0.86191954999425424, 0.86692047674867767]
Since each additional tree allows the model to see more data, this approach can make additional trees more useful.
m = RandomForestRegressor(n_estimators=40, n_jobs=-1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
[0.2317315086850927, 0.26334275954117264, 0.89225792718146846, 0.87615150359885019, 0.88097587673696554]
Tree building parameters¶
We revert to using a full bootstrap sample in order to show the impact of other over-fitting avoidance methods.
reset_rf_samples()
Let's get a baseline for this full set to compare to.
def dectree_max_depth(tree):
children_left = tree.children_left
children_right = tree.children_right
def walk(node_id):
if (children_left[node_id] != children_right[node_id]):
left_max = 1 + walk(children_left[node_id])
right_max = 1 + walk(children_right[node_id])
return max(left_max, right_max)
else: # leaf
return 1
root_node_id = 0
return walk(root_node_id)
m = RandomForestRegressor(n_estimators=40, n_jobs=-1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
[0.07828713008286803, 0.23818558990341943, 0.9871909898049919, 0.8986837887808402, 0.9085077721150765]
t=m.estimators_[0].tree_
dectree_max_depth(t)
45
m = RandomForestRegressor(n_estimators=40, min_samples_leaf=5, n_jobs=-1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
[0.14073508031497292, 0.23337403295759937, 0.9586057939941005, 0.9027357960501001, 0.9068706269691232]
t=m.estimators_[0].tree_
dectree_max_depth(t)
35
Another way to reduce over-fitting is to grow our trees less deeply. We do this by specifying (with min_samples_leaf) that we require some minimum number of rows in every leaf node. This has two benefits:
- There are less decision rules for each leaf node; simpler models should generalize better
- The predictions are made by averaging more rows in the leaf node, resulting in less volatility
m = RandomForestRegressor(n_estimators=40, min_samples_leaf=3, n_jobs=-1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
[0.11595869956476182, 0.23427349924625201, 0.97209195463880227, 0.90198460308551043, 0.90843297242839738]
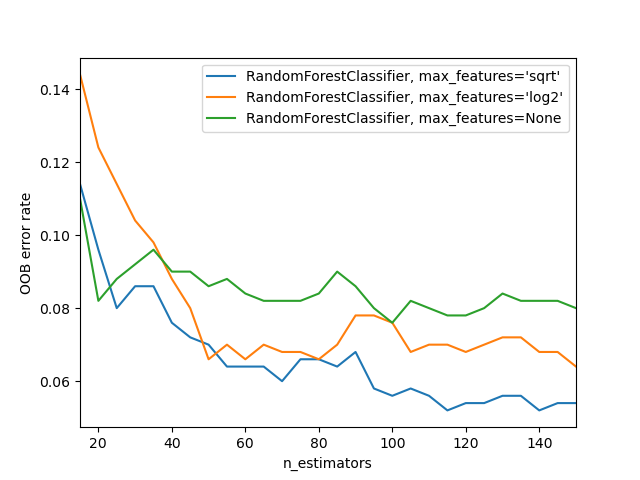
We can also increase the amount of variation amongst the trees by not only use a sample of rows for each tree, but to also using a sample of columns for each split. We do this by specifying max_features, which is the proportion of features to randomly select from at each split.
- None
- 0.5
- 'sqrt'
- 1, 3, 5, 10, 25, 100
m = RandomForestRegressor(n_estimators=40, min_samples_leaf=3, max_features=0.5, n_jobs=-1, oob_score=True)
m.fit(X_train, y_train)
print_score(m)
[0.11926975747908228, 0.22869111042050522, 0.97026995966445684, 0.9066000722129437, 0.91144914977164715]
We can't compare our results directly with the Kaggle competition, since it used a different validation set (and we can no longer to submit to this competition) - but we can at least see that we're getting similar results to the winners based on the dataset we have.
The sklearn docs show an example of different max_features methods with increasing numbers of trees - as you see, using a subset of features on each split requires using more trees, but results in better models: