Randomized SVD demos¶
Part 1: Random Projections (with word vectors)¶
The purpose of this section is to illustrate the idea of random projections preserving structure with the concrete example of word vectors!
To use language in machine learning (for instance, how Skype translator translates between languages, or how Gmail Smart Reply automatically suggests possible responses for your emails), we need to represent words as vectors.
We can represent words as 100 dimensional vectors, using Google's Word2Vec or Stanford's GloVe. For example, here is the word "python" as a vector in GloVe:
vecs[wordidx['python']]
array([ 0.2493, 0.6832, -0.0447, -1.3842, -0.0073, 0.651 , -0.3396,
-0.1979, -0.3392, 0.2669, -0.0331, 0.1592, 0.8955, 0.54 ,
-0.5582, 0.4624, 0.3672, 0.1889, 0.8319, 0.8142, -0.1183,
-0.5346, 0.2416, -0.0389, 1.1907, 0.7935, -0.1231, 0.6642,
-0.7762, -0.4571, -1.054 , -0.2056, -0.133 , 0.1224, 0.8846,
1.024 , 0.3229, 0.821 , -0.0694, 0.0242, -0.5142, 0.8727,
0.2576, 0.9153, -0.6422, 0.0412, -0.6021, 0.5463, 0.6608,
0.198 , -1.1393, 0.7951, 0.4597, -0.1846, -0.6413, -0.2493,
-0.4019, -0.5079, 0.8058, 0.5336, 0.5273, 0.3925, -0.2988,
0.0096, 0.9995, -0.0613, 0.7194, 0.329 , -0.0528, 0.6714,
-0.8025, -0.2579, 0.4961, 0.4808, -0.684 , -0.0122, 0.0482,
0.2946, 0.2061, 0.3356, -0.6417, -0.6471, 0.1338, -0.1257,
-0.4638, 1.3878, 0.9564, -0.0679, -0.0017, 0.5296, 0.4567,
0.6104, -0.1151, 0.4263, 0.1734, -0.7995, -0.245 , -0.6089,
-0.3847, -0.4797], dtype=float32)
Goal: Use randomness to reduce this from 100 dimensions to 20. Check that similar words are still grouped together.
More info: If you are interested in word embeddings and want more detail, I gave a longer workshop about them available here (with code demo).
Style note: I use collapsible headings and jupyter themes
Loading the data¶
import pickle
import numpy as np
import re
import json
np.set_printoptions(precision=4, suppress=True)
The dataset is available at http://files.fast.ai/models/glove/6B.100d.tgz To download and unzip the files from the command line, you can run:
wget http://files.fast.ai/models/glove_50_glove_100.tgz
tar xvzf glove_50_glove_100.tgz
You will need to update the path below to be accurate for where you are storing the data.
path = "../data/"
vecs = np.load(path + "glove_vectors_100d.npy")
with open(path + "words.txt") as f:
content = f.readlines()
words = [x.strip() for x in content]
wordidx = json.load(open(path + "wordsidx.txt"))
What our data looks like¶
We have a long list of words:
len(words)
400000
words[:10]
['the', ',', '.', 'of', 'to', 'and', 'in', 'a', '"', "'s"]
words[600:610]
['together', 'congress', 'index', 'australia', 'results', 'hard', 'hours', 'land', 'action', 'higher']
wordidx allows us to look up a word in order to find out it's index:
wordidx['python']
20019
words[20019]
'python'
Words as vectors¶
The word "python" is represented by the 100 dimensional vector:
vecs[wordidx['python']]
array([ 0.2493, 0.6832, -0.0447, -1.3842, -0.0073, 0.651 , -0.3396,
-0.1979, -0.3392, 0.2669, -0.0331, 0.1592, 0.8955, 0.54 ,
-0.5582, 0.4624, 0.3672, 0.1889, 0.8319, 0.8142, -0.1183,
-0.5346, 0.2416, -0.0389, 1.1907, 0.7935, -0.1231, 0.6642,
-0.7762, -0.4571, -1.054 , -0.2056, -0.133 , 0.1224, 0.8846,
1.024 , 0.3229, 0.821 , -0.0694, 0.0242, -0.5142, 0.8727,
0.2576, 0.9153, -0.6422, 0.0412, -0.6021, 0.5463, 0.6608,
0.198 , -1.1393, 0.7951, 0.4597, -0.1846, -0.6413, -0.2493,
-0.4019, -0.5079, 0.8058, 0.5336, 0.5273, 0.3925, -0.2988,
0.0096, 0.9995, -0.0613, 0.7194, 0.329 , -0.0528, 0.6714,
-0.8025, -0.2579, 0.4961, 0.4808, -0.684 , -0.0122, 0.0482,
0.2946, 0.2061, 0.3356, -0.6417, -0.6471, 0.1338, -0.1257,
-0.4638, 1.3878, 0.9564, -0.0679, -0.0017, 0.5296, 0.4567,
0.6104, -0.1151, 0.4263, 0.1734, -0.7995, -0.245 , -0.6089,
-0.3847, -0.4797], dtype=float32)
This lets us do some useful calculations. For instance, we can see how far apart two words are using a distance metric:
from scipy.spatial.distance import cosine as dist
Smaller numbers mean two words are closer together, larger numbers mean they are further apart.
The distance between similar words is low:
dist(vecs[wordidx["puppy"]], vecs[wordidx["dog"]])
0.27636240676695256
dist(vecs[wordidx["queen"]], vecs[wordidx["princess"]])
0.20527545040329642
And the distance between unrelated words is high:
dist(vecs[wordidx["celebrity"]], vecs[wordidx["dusty"]])
0.98835787578057777
dist(vecs[wordidx["avalanche"]], vecs[wordidx["antique"]])
0.96211070091611983
Bias¶
There is a lot of opportunity for bias:
dist(vecs[wordidx["man"]], vecs[wordidx["genius"]])
0.50985148631697985
dist(vecs[wordidx["woman"]], vecs[wordidx["genius"]])
0.6897833082810727
I just checked the distance between pairs of words, because this is a quick and simple way to illustrate the concept. It is also a very noisy approach, and researchers approach this problem in more systematic ways.
I talk about bias in much greater depth in this workshop.
Visualizations¶
Let's visualize some words!
We will use Plotly, a Python library to make interactive graphs (note: everything below is done without creating an account, with the free, offline version of Plotly).
Methods¶
import plotly
import plotly.graph_objs as go
from IPython.display import IFrame
def plotly_3d(Y, cat_labels, filename="temp-plot.html"):
trace_dict = {}
for i, label in enumerate(cat_labels):
trace_dict[i] = go.Scatter3d(
x=Y[i*5:(i+1)*5, 0],
y=Y[i*5:(i+1)*5, 1],
z=Y[i*5:(i+1)*5, 2],
mode='markers',
marker=dict(
size=8,
line=dict(
color='rgba('+ str(i*40) + ',' + str(i*40) + ',' + str(i*40) + ', 0.14)',
width=0.5
),
opacity=0.8
),
text = my_words[i*5:(i+1)*5],
name = label
)
data = [item for item in trace_dict.values()]
layout = go.Layout(
margin=dict(
l=0,
r=0,
b=0,
t=0
)
)
plotly.offline.plot({
"data": data,
"layout": layout,
}, filename=filename)
def plotly_2d(Y, cat_labels, filename="temp-plot.html"):
trace_dict = {}
for i, label in enumerate(cat_labels):
trace_dict[i] = go.Scatter(
x=Y[i*5:(i+1)*5, 0],
y=Y[i*5:(i+1)*5, 1],
mode='markers',
marker=dict(
size=8,
line=dict(
color='rgba('+ str(i*40) + ',' + str(i*40) + ',' + str(i*40) + ', 0.14)',
width=0.5
),
opacity=0.8
),
text = my_words[i*5:(i+1)*5],
name = label
)
data = [item for item in trace_dict.values()]
layout = go.Layout(
margin=dict(
l=0,
r=0,
b=0,
t=0
)
)
plotly.offline.plot({
"data": data,
"layout": layout
}, filename=filename)
This method will pick out the 3 dimensions that best separate our categories from one another (stored in dist_btwn_cats), while minimizing the distance of the words within a given category (stored in dist_within_cats).
def get_components(data, categories, word_indices):
num_components = 30
pca = decomposition.PCA(n_components=num_components).fit(data.T)
all_components = pca.components_
centroids = {}
print(all_components.shape)
for i, category in enumerate(categories):
cen = np.mean(all_components[:, i*5:(i+1)*5], axis = 1)
dist_within_cats = np.sum(np.abs(np.expand_dims(cen, axis=1) - all_components[:, i*5:(i+1)*5]), axis=1)
centroids[category] = cen
dist_btwn_cats = np.zeros(num_components)
for category1, averages1 in centroids.items():
for category2, averages2 in centroids.items():
dist_btwn_cats += abs(averages1 - averages2)
clusterness = dist_btwn_cats / dist_within_cats
comp_indices = np.argpartition(clusterness, -3)[-3:]
return all_components[comp_indices]
Preparing the Data¶
Let's plot words from a few different categories:
my_words = [
"maggot", "flea", "tarantula", "bedbug", "mosquito",
"violin", "cello", "flute", "harp", "mandolin",
"joy", "love", "peace", "pleasure", "wonderful",
"agony", "terrible", "horrible", "nasty", "failure",
"physics", "chemistry", "science", "technology", "engineering",
"poetry", "art", "literature", "dance", "symphony",
]
categories = [
"bugs", "music",
"pleasant", "unpleasant",
"science", "arts"
]
Again, we need to look up the indices of our words using the wordidx dictionary:
my_word_indices = np.array([wordidx[word] for word in my_words])
vecs[my_word_indices].shape
(30, 100)
Now, we will make a set combining our words with the first 10,000 words in our entire set of words (some of the words will already be in there), and create a matrix of their embeddings.
embeddings = np.concatenate((vecs[my_word_indices], vecs[:10000,:]), axis=0); embeddings.shape
(10030, 100)
Viewing the words in 3D¶
The words are in 100 dimensions and we need a way to visualize them in 3D.
We will use Principal Component Analysis (PCA), a widely used technique with many applications, including visualizing high-dimensional data sets in a lower dimension!
PCA¶
from collections import defaultdict
from sklearn import decomposition
components = get_components(embeddings, categories, my_word_indices)
plotly_3d(components.T[:len(my_words),:], categories, "pca.html")
(30, 10030)
IFrame('pca.html', width=600, height=400)
Random Projections¶
Johnson-Lindenstrauss Lemma: (from wikipedia) a small set of points in a high-dimensional space can be embedded into a space of much lower dimension in such a way that distances between the points are nearly preserved (proof uses random projections).
It is useful to be able to reduce dimensionality of data in a way that preserves distances. The Johnson–Lindenstrauss lemma is a classic result of this type.
embeddings.shape
(10030, 100)
rand_proj = embeddings @ np.random.normal(size=(embeddings.shape[1], 40)); rand_proj.shape
(10030, 40)
# pca = decomposition.PCA(n_components=3).fit(rand_proj.T)
# components = pca.components_
components = get_components(rand_proj, categories, my_word_indices)
plotly_3d(components.T[:len(my_words),:], categories, "pca-rand-proj.html")
(30, 10030)
IFrame('pca-rand-proj.html', width=600, height=400)
Part 2: Background Removal¶
Our goal today: 
Load and Format the Data¶
Let's use the real video 003 dataset from BMC 2012 Background Models Challenge Dataset
Import needed libraries:
import imageio
imageio.plugins.ffmpeg.download()
import moviepy.editor as mpe
import numpy as np
import scipy
%matplotlib inline
import matplotlib.pyplot as plt
video = mpe.VideoFileClip("videos/Video_003.avi")
video.subclip(0,50).ipython_display(width=300)
100%|█████████▉| 350/351 [00:00<00:00, 1097.29it/s]
video.duration
113.57
Helper Methods¶
def create_data_matrix_from_video(clip, fps=5, scale=50):
return np.vstack([scipy.misc.imresize(rgb2gray(clip.get_frame(i/float(fps))).astype(int),
scale).flatten() for i in range(fps * int(clip.duration))]).T
def rgb2gray(rgb):
return np.dot(rgb[...,:3], [0.299, 0.587, 0.114])
Format the Data¶
An image from 1 moment in time is 120 pixels by 160 pixels (when scaled). We can unroll that picture into a single tall column. So instead of having a 2D picture that is $120 \times 160$, we have a $1 \times 19,200$ column
This isn't very human-readable, but it's handy because it lets us stack the images from different times on top of one another, to put a video all into 1 matrix. If we took the video image every hundredth of a second for 100 seconds (so 10,000 different images, each from a different point in time), we'd have a $10,000 \times 19,200$ matrix, representing the video!
scale = 0.50 # Adjust scale to change resolution of image
dims = (int(240 * scale), int(320 * scale))
fps = 60 # frames per second
M = create_data_matrix_from_video(video.subclip(0,100), fps, scale)
# M = np.load("med_res_surveillance_matrix.npy")
print(dims, M.shape)
(120, 160) (19200, 6000)
plt.imshow(np.reshape(M[:,140], dims), cmap='gray');
Since create_data_from_matrix is somewhat slow, we will save our matrix. In general, whenever you have slow pre-processing steps, it's a good idea to save the results for future use.
np.save("med_res_surveillance_matrix_60fps.npy", M)
plt.figure(figsize=(12, 12))
plt.imshow(M, cmap='gray')
<matplotlib.image.AxesImage at 0x7f601f315fd0>
Questions: What are those wavy black lines? What are the horizontal lines?
Part 2 continued: SVD¶
Intro to SVD¶
Applications of SVD:
- Principal Component Analysis
- Data compression
- Pseudo-inverse
- Collaborative Filtering
- Topic Modeling
- Background Removal
- Removing Corrupted Data
“a convenient way for breaking a matrix into simpler, meaningful pieces we care about” – David Austin
“the most important linear algebra concept I don’t remember learning” - Daniel Lemire
U, s, V = np.linalg.svd(M, full_matrices=False)
This is really slow, so you may want to save your result to use in the future.
np.save("U.npy", U)
np.save("s.npy", s)
np.save("V.npy", V)
In the future, you can just load what you've saved:
U = np.load("U.npy")
s = np.load("s.npy")
V = np.load("V.npy")
What do $U$, $S$, and $V$ look like?
U.shape, s.shape, V.shape
((19200, 6000), (6000,), (6000, 6000))
Check that they are a decomposition of M
reconstructed_matrix = U @ np.diag(s) @ V
np.allclose(M, reconstructed_matrix)
True
They are! :-)
np.set_printoptions(suppress=True, precision=0)
s is the diagonal of a diagonal matrix
np.diag(s[:6])
array([[ 1341720., 0., 0., 0., 0., 0.],
[ 0., 40231., 0., 0., 0., 0.],
[ 0., 0., 35092., 0., 0., 0.],
[ 0., 0., 0., 30997., 0., 0.],
[ 0., 0., 0., 0., 28220., 0.],
[ 0., 0., 0., 0., 0., 27196.]])
Do you see anything about the order for $s$?
s[0:2000:50]
array([ 1341720., 10528., 6162., 4235., 3174., 2548.,
2138., 1813., 1558., 1346., 1163., 1001.,
841., 666., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0.,
0., 0., 0., 0.])
len(s)
6000
s[700]
3.2309523518534773e-10
np.set_printoptions(suppress=True, precision=4)
$U$ is a giant matrix, so let's just look at a tiny bit of it:
U[:5,:5]
array([[-0.0083, -0.0009, -0.0007, 0.003 , -0.0002],
[-0.0083, -0.0013, -0.0005, 0.0034, -0.0001],
[-0.0084, -0.0012, 0.0002, 0.0045, -0.0003],
[-0.0085, -0.0011, 0.0001, 0.0044, -0. ],
[-0.0086, -0.0013, -0.0002, 0.004 , 0.0001]])
Finding the background¶
U.shape, s.shape, V.shape
((19200, 6000), (6000,), (6000, 6000))
low_rank = np.expand_dims(U[:,0], 1) * s[0] * np.expand_dims(V[0,:], 0)
plt.figure(figsize=(12, 12))
plt.imshow(low_rank, cmap='gray')
<matplotlib.image.AxesImage at 0x7f1cc3e2c9e8>
plt.imshow(np.reshape(low_rank[:,0], dims), cmap='gray');
How do we get the people from here?
plt.imshow(np.reshape(M[:,0] - low_rank[:,0], dims), cmap='gray');
High-resolution version
plt.imshow(np.reshape(M[:,140] - low_rank[:,140], dims), cmap='gray');
Make Video¶
I was inspired by the work of fast.ai student Samir Moussa to make videos of the people.
from moviepy.video.io.bindings import mplfig_to_npimage
def make_video(matrix, dims, filename):
mat_reshaped = np.reshape(matrix, (dims[0], dims[1], -1))
fig, ax = plt.subplots()
def make_frame(t):
ax.clear()
ax.imshow(mat_reshaped[...,int(t*fps)])
return mplfig_to_npimage(fig)
animation = mpe.VideoClip(make_frame, duration=int(10))
animation.write_videofile('videos/' + filename + '.mp4', fps=fps)
make_video(M - low_rank, dims, "figures2")
[MoviePy] >>>> Building video videos/figures2.mp4 [MoviePy] Writing video videos/figures2.mp4
100%|█████████▉| 600/601 [00:39<00:00, 15.22it/s]
[MoviePy] Done. [MoviePy] >>>> Video ready: videos/figures2.mp4
mpe.VideoFileClip("videos/figures2.mp4").subclip(0,10).ipython_display(width=300)
100%|█████████▉| 600/601 [00:00<00:00, 858.48it/s]
Speed of SVD for different size matrices¶
import timeit
import pandas as pd
m_array = np.array([100, int(1e3), int(1e4)])
n_array = np.array([100, int(1e3), int(1e4)])
index = pd.MultiIndex.from_product([m_array, n_array], names=['# rows', '# cols'])
pd.options.display.float_format = '{:,.3f}'.format
df = pd.DataFrame(index=m_array, columns=n_array)
# %%prun
for m in m_array:
for n in n_array:
A = np.random.uniform(-40,40,[m,n])
t = timeit.timeit('np.linalg.svd(A, full_matrices=False)', number=3, globals=globals())
df.set_value(m, n, t)
df/3
| 100 | 1000 | 10000 | |
|---|---|---|---|
| 100 | 0.006 | 0.009 | 0.043 |
| 1000 | 0.004 | 0.259 | 0.992 |
| 10000 | 0.019 | 0.984 | 218.726 |
2 Backgrounds in 1 Video¶
We'll now use real video 008 dataset from BMC 2012 Background Models Challenge Dataset, in addition to video 003 that we used above.
video2 = mpe.VideoFileClip("videos/Video_008.avi")
from moviepy.editor import concatenate_videoclips
concat_video = concatenate_videoclips([video2.subclip(0,60), video.subclip(0,100)])
concat_video.write_videofile("concatenated_video.mp4")
[MoviePy] >>>> Building video concatenated_video.mp4 [MoviePy] Writing video concatenated_video.mp4
100%|█████████▉| 1600/1601 [00:02<00:00, 751.90it/s]
[MoviePy] Done. [MoviePy] >>>> Video ready: concatenated_video.mp4
concat_video.ipython_display(width=300)
100%|█████████▉| 500/501 [00:00<00:00, 563.14it/s]
scale = 0.5 # Adjust scale to change resolution of image
dims = (int(240 * scale), int(320 * scale))
N = create_data_matrix_from_video(concat_video, fps, scale)
# N = np.load("low_res_traffic_matrix.npy")
np.save("med_res_concat_video.npy", N)
N.shape
(19200, 10000)
plt.imshow(np.reshape(N[:,200], dims), cmap='gray');
U_concat, s_concat, V_concat = np.linalg.svd(N, full_matrices=False)
This is slow, so you may want to save your result to use in the future.
np.save("U_concat.npy", U_concat)
np.save("s_concat.npy", s_concat)
np.save("V_concat.npy", V_concat)
In the future, you can just load what you've saved:
U_concat = np.load("U_concat.npy")
s_concat = np.load("s_concat.npy")
V_concat = np.load("V_concat.npy")
low_rank = U_concat[:,:10] @ np.diag(s_concat[:10]) @ V_concat[:10,:]
The top few components of U:
plt.imshow(np.reshape(U_concat[:, 1], dims), cmap='gray')
<matplotlib.image.AxesImage at 0x7f1cc3587780>
plt.imshow(np.reshape(U_concat[:, 2], dims), cmap='gray')
<matplotlib.image.AxesImage at 0x7f1cc33e1a90>
plt.imshow(np.reshape(U_concat[:, 3], dims), cmap='gray')
<matplotlib.image.AxesImage at 0x7f1cc3a46438>
Background removal:
plt.imshow(np.reshape((N - low_rank)[:, -40], dims), cmap='gray')
<matplotlib.image.AxesImage at 0x7f1cc3f8dda0>
plt.imshow(np.reshape((N - low_rank)[:, 240], dims), cmap='gray')
<matplotlib.image.AxesImage at 0x7f1cc3c23b70>
Aside about data compression¶
Suppose we take 700 singular values (remember, there are 10000 singular values total)
s[0:1000:50]
array([ 1341719.6552, 10527.5148, 6162.0638, 4234.9367,
3174.0389, 2548.4273, 2138.1887, 1812.9873,
1557.7163, 1345.805 , 1163.2866, 1000.5186,
841.4604, 665.7271, 0. , 0. ,
0. , 0. , 0. , 0. ])
k = 700
compressed_M = U[:,:k] @ np.diag(s[:k]) @ V[:k,:]
plt.figure(figsize=(12, 12))
plt.imshow(compressed_M, cmap='gray')
<matplotlib.image.AxesImage at 0x7fefa0076ac8>
plt.imshow(np.reshape(compressed_M[:,140], dims), cmap='gray');
np.allclose(compressed_M, M)
True
np.linalg.norm(M - compressed_M)
2.864899899979104e-09
U[:,:k].shape, s[:k].shape, V[:k,:].shape
((19200, 700), (700,), (700, 6000))
space saved = data in U, s, V for 700 singular values / original matrix
((19200 + 1 + 6000) * 700) / (19200 * 6000)
0.1531310763888889
We only need to store 15.3% as much data and can keep the accuracy to 1e-5! That's great!
Now back to our background removal problem:¶
That's pretty neat!!! but...¶
Downside: this was really slow (also, we threw away a lot of our calculation)
%time u, s, v = np.linalg.svd(M, full_matrices=False)
CPU times: user 5min 38s, sys: 1.53 s, total: 5min 40s Wall time: 57.1 s
M.shape
(19200, 6000)
The runtime complexity for SVD is $\mathcal{O}(\text{min}(m^2 n,\; m n^2))$
Let's create a smaller version of M¶
Idea: Let's use a smaller matrix!
We haven't found a better general SVD method, we'll just use the method we have on a smaller matrix.
def randomized_svd(M, k=10):
m, n = M.shape
transpose = False
if m < n:
transpose = True
M = M.T
rand_matrix = np.random.normal(size=(M.shape[1], k)) # short side by k
Q, _ = np.linalg.qr(M @ rand_matrix, mode='reduced') # long side by k
smaller_matrix = Q.T @ M # k by short side
U_hat, s, V = np.linalg.svd(smaller_matrix, full_matrices=False)
U = Q @ U_hat
if transpose:
return V.T, s.T, U.T
else:
return U, s, V
%time u, s, v = randomized_svd(M, 10)
CPU times: user 3.06 s, sys: 268 ms, total: 3.33 s Wall time: 789 ms
U_rand, s_rand, V_rand = randomized_svd(M, 10)
low_rank = np.expand_dims(U_rand[:,0], 1) * s_rand[0] * np.expand_dims(V_rand[0,:], 0)
plt.imshow(np.reshape(low_rank[:,0], dims), cmap='gray');
How do we get the people from here?
plt.imshow(np.reshape(M[:,0] - low_rank[:,0], dims), cmap='gray');
What this method is doing¶
rand_matrix = np.random.normal(size=(M.shape[1], 10))
rand_matrix.shape
(6000, 10)
plt.imshow(np.reshape(rand_matrix[:4900,0], (70,70)), cmap='gray');
temp = M @ rand_matrix; temp.shape
(19200, 10)
plt.imshow(np.reshape(temp[:,0], dims), cmap='gray');
plt.imshow(np.reshape(temp[:,1], dims), cmap='gray');
Q, _ = np.linalg.qr(M @ rand_matrix, mode='reduced'); Q.shape
(19200, 10)
np.dot(Q[:,0], Q[:,1])
-3.8163916471489756e-17
plt.imshow(np.reshape(Q[:,0], dims), cmap='gray');
plt.imshow(np.reshape(Q[:,1], dims), cmap='gray');
smaller_matrix = Q.T @ M; smaller_matrix.shape
(10, 6000)
U_hat, s, V = np.linalg.svd(smaller_matrix, full_matrices=False)
U = Q @ U_hat
plt.imshow(np.reshape(U[:,0], dims), cmap='gray');
reconstructed_small_M = U @ np.diag(s) @ V
And the people:
plt.imshow(np.reshape(M[:,0] - reconstructed_small_M[:,0], dims), cmap='gray');
Timing Comparison¶
from sklearn import decomposition
import fbpca
Full SVD:
%time u, s, v = np.linalg.svd(M, full_matrices=False)
CPU times: user 5min 38s, sys: 1.53 s, total: 5min 40s Wall time: 57.1 s
Our (overly simplified) randomized_svd from above:
%time u, s, v = randomized_svd(M, 10)
CPU times: user 2.37 s, sys: 160 ms, total: 2.53 s Wall time: 641 ms
Scikit learn:
%time u, s, v = decomposition.randomized_svd(M, 10)
CPU times: user 19.2 s, sys: 1.44 s, total: 20.7 s Wall time: 3.67 s
Randomized SVD from Facebook's library fbpca:
%time u, s, v = fbpca.pca(M, 10)
CPU times: user 7.28 s, sys: 424 ms, total: 7.7 s Wall time: 1.37 s
I would choose fbpca, since it's faster than sklearn but more robust and more accurate than our simple implementation.
Here are some results from Facebook Research:
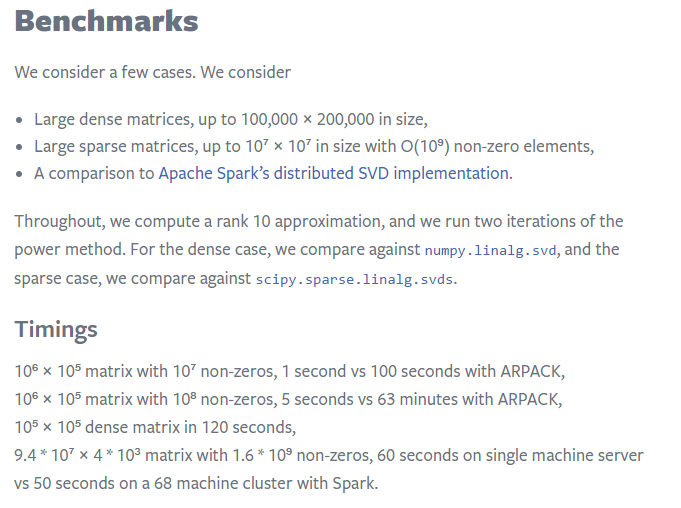
Time and Accuracy vary with k¶
import timeit
import pandas as pd
U_rand, s_rand, V_rand = fbpca.pca(M, 700, raw=True)
reconstructed = U_rand @ np.diag(s_rand) @ V_rand
np.linalg.norm(M - reconstructed)
1.1065914828881536e-07
plt.imshow(np.reshape(reconstructed[:,140], dims), cmap='gray');
pd.options.display.float_format = '{:,.2f}'.format
k_values = np.arange(100,1000,100)
df_rand = pd.DataFrame(index=["time", "error"], columns=k_values)
# df_rand = pd.read_pickle("svd_df")
for k in k_values:
U_rand, s_rand, V_rand = fbpca.pca(M, k, raw=True)
reconstructed = U_rand @ np.diag(s_rand) @ V_rand
df_rand.set_value("error", k, np.linalg.norm(M - reconstructed))
t = timeit.timeit('fbpca.pca(M, k)', number=3, globals=globals())
df_rand.set_value("time", k, t/3)
df_rand.to_pickle("df_rand")
df_rand
| 100 | 200 | 300 | 400 | 500 | 600 | 700 | 800 | 900 | 1000 | |
|---|---|---|---|---|---|---|---|---|---|---|
| time | 2.07 | 2.57 | 3.45 | 6.44 | 7.99 | 9.02 | 10.24 | 11.70 | 13.30 | 10.87 |
| error | 58,997.27 | 37,539.54 | 26,569.89 | 18,769.37 | 12,559.34 | 6,936.17 | 0.00 | 0.00 | 0.00 | 0.00 |
df = pd.DataFrame(index=["error"], columns=k_values)
for k in k_values:
reconstructed = U[:,:k] @ np.diag(s[:k]) @ V[:k,:]
df.set_value("error", k, np.linalg.norm(M - reconstructed))
df.to_pickle("df")
fig, ax1 = plt.subplots()
ax1.plot(df.columns, df_rand.loc["time"].values, 'b-', label="randomized SVD time")
ax1.plot(df.columns, np.tile([57], 9), 'g-', label="SVD time")
ax1.set_xlabel('k: # of singular values')
# Make the y-axis label, ticks and tick labels match the line color.
ax1.set_ylabel('time', color='b')
ax1.tick_params('y', colors='b')
ax1.legend(loc = 0)
ax2 = ax1.twinx()
ax2.plot(df.columns, df_rand.loc["error"].values, 'r--', label="randomized SVD error")
ax2.plot(df.columns, df.loc["error"].values, 'm--', label="SVD error")
ax2.set_ylabel('error', color='r')
ax2.tick_params('y', colors='r')
ax2.legend(loc=1)
#fig.tight_layout()
plt.show()
Math Details¶
Process behind Randomized SVD¶
Here is a process to calculate a truncated SVD, described in Finding Structure with Randomness: Probabilistic Algorithms for Constructing Approximate Matrix Decompositions and summarized in this blog post:
1. Compute an approximation to the range of $A$. That is, we want $Q$ with $r$ orthonormal columns such that $$A \approx QQ^TA$$
2. Construct $B = Q^T A$, which is small ($r\times n$)
3. Compute the SVD of $B$ by standard methods (fast since $B$ is smaller than $A$), $B = S\,\Sigma V^T$
4. Since $$ A \approx Q Q^T A = Q (S\,\Sigma V^T)$$ if we set $U = QS$, then we have a low rank approximation $A \approx U \Sigma V^T$.
So how do we find $Q$ (in step 1)?¶
To estimate the range of $A$, we can just take a bunch of random vectors $w_i$, evaluate the subspace formed by $Aw_i$. We can form a matrix $W$ with the $w_i$ as it's columns. Now, we take the QR decomposition of $AW = QR$, then the columns of $Q$ form an orthonormal basis for $AW$, which is the range of $A$.
Since the matrix $AW$ of the product has far more rows than columns and therefore, approximately, orthonormal columns. This is simple probability - with lots of rows, and few columns, it's unlikely that the columns are linearly dependent.
Why M ~ Q Q.T M¶
We are trying to find a matrix Q such that $M \approx Q Q^T M$. We are interested in the range of $M$, let's call this $MX$. $Q$ has orthonormal columns so $Q^TQ = I$ (but $QQ^T$ isn't $I$, since $Q$ is rectangular)
$$ QR = MX $$$$ QQ^TQR = QQ^TMX $$$$ QR = QQ^TMX $$so... $$ MX = QQ^TMX $$
If $X$ is the identity, we'd be done (but then $X$ would be too big, and we wouldn't get the speed up we're looking for). In our problem, $X$ is just a small random matrix. The Johnson-Lindenstrauss Lemma provides some justification of why this works.
The QR Decomposition¶
We will be learning about the QR decomposition in depth later on. For now, you just need to know that $A = QR$, where $Q$ consists of orthonormal columns, and $R$ is upper triangular. Trefethen says that the QR decomposition is the most important idea in numerical linear algebra! We will definitely be returning to it.
How should we choose $r$?¶
Suppose our matrix has 100 columns, and we want 5 columns in U and V. To be safe, we should project our matrix onto an orthogonal basis with a few more rows and columns than 5 (let's use 15). At the end, we will just grab the first 5 columns of U and V
So even although our projection was only approximate, by making it a bit bigger than we need, we can make up for the loss of accuracy (since we're only taking a subset later).
How this is different from random mean¶
test = M @ np.random.normal(size=(M.shape[1], 2)); test.shape
(4800, 2)
Random mean:
plt.imshow(np.reshape(test[:,0], dims), cmap='gray');
Mean image:
plt.imshow(np.reshape(M.mean(axis=1), dims), cmap='gray')
<matplotlib.image.AxesImage at 0x7f83f4093fd0>
plt.imshow(np.reshape(test[:,1], dims), cmap='gray');
ut, st, vt = np.linalg.svd(test, full_matrices=False)
plt.imshow(np.reshape(smaller_matrix[0,:], dims), cmap='gray');
plt.imshow(np.reshape(smaller_matrix[1,:], dims), cmap='gray');
plt.imshow(np.reshape(M[:,140], dims), cmap='gray');