stackview + pyclesperanto_prototype demo¶
Interactive image stack viewing in jupyter notebooks based on ipycanvas and ipywidgets.
In this notebook we demonstate its usage on google colab in combination with pyclesperanto_prototype.
Run this notebook in google colab
Installation¶
After starting a new session on google colab, you need to install stackview and pyclesperanto-prototype first. After this, you need to restart the kernel. Use the opportunity to click the menu Runtime > Change runtime type and activate the GPU.
Also note: The make stackview work in google colab, you need to install ipycanvas==0.11 (read why).
!pip install pyclesperanto_prototype stackview ipycanvas==0.11
Requirement already satisfied: pyclesperanto_prototype in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (0.24.0) Requirement already satisfied: stackview in c:\structure\code\stackview (0.6.1) Requirement already satisfied: ipycanvas==0.11 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (0.11.0) Requirement already satisfied: numpy in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipycanvas==0.11) (1.23.5) Requirement already satisfied: ipywidgets>=7.6.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipycanvas==0.11) (8.0.6) Requirement already satisfied: pillow>=6.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipycanvas==0.11) (9.2.0) Requirement already satisfied: pyopencl in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyclesperanto_prototype) (2022.3.1) Requirement already satisfied: toolz in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyclesperanto_prototype) (0.12.0) Requirement already satisfied: matplotlib in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyclesperanto_prototype) (3.7.1) Requirement already satisfied: scikit-image!=0.20.0,>=0.18.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyclesperanto_prototype) (0.19.3) Requirement already satisfied: transforms3d in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyclesperanto_prototype) (0.4.1) Requirement already satisfied: ipyevents in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from stackview) (2.0.1) Requirement already satisfied: ipykernel in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from stackview) (6.22.0) Requirement already satisfied: widgetsnbextension~=4.0.7 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipywidgets>=7.6.0->ipycanvas==0.11) (4.0.7) Requirement already satisfied: ipython>=6.1.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipywidgets>=7.6.0->ipycanvas==0.11) (8.12.0) Requirement already satisfied: jupyterlab-widgets~=3.0.7 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipywidgets>=7.6.0->ipycanvas==0.11) (3.0.7) Requirement already satisfied: traitlets>=4.3.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipywidgets>=7.6.0->ipycanvas==0.11) (5.9.0) Requirement already satisfied: nest-asyncio in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (1.5.6) Requirement already satisfied: comm>=0.1.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (0.1.3) Requirement already satisfied: debugpy>=1.6.5 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (1.6.7) Requirement already satisfied: matplotlib-inline>=0.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (0.1.6) Requirement already satisfied: tornado>=6.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (6.2) Requirement already satisfied: psutil in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (5.9.4) Requirement already satisfied: jupyter-client>=6.1.12 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (8.2.0) Requirement already satisfied: pyzmq>=20 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (25.0.2) Requirement already satisfied: packaging in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (23.1) Requirement already satisfied: jupyter-core!=5.0.*,>=4.12 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipykernel->stackview) (5.3.0) Requirement already satisfied: scipy>=1.4.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from scikit-image!=0.20.0,>=0.18.0->pyclesperanto_prototype) (1.10.1) Requirement already satisfied: imageio>=2.4.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from scikit-image!=0.20.0,>=0.18.0->pyclesperanto_prototype) (2.27.0) Requirement already satisfied: networkx>=2.2 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from scikit-image!=0.20.0,>=0.18.0->pyclesperanto_prototype) (3.1) Requirement already satisfied: tifffile>=2019.7.26 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from scikit-image!=0.20.0,>=0.18.0->pyclesperanto_prototype) (2022.10.10) Requirement already satisfied: PyWavelets>=1.1.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from scikit-image!=0.20.0,>=0.18.0->pyclesperanto_prototype) (1.4.1) Requirement already satisfied: python-dateutil>=2.7 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (2.8.2) Requirement already satisfied: importlib-resources>=3.2.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (5.12.0) Requirement already satisfied: pyparsing>=2.3.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (3.0.9) Requirement already satisfied: contourpy>=1.0.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (1.0.7) Requirement already satisfied: cycler>=0.10 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (0.11.0) Requirement already satisfied: fonttools>=4.22.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (4.39.3) Requirement already satisfied: kiwisolver>=1.0.1 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from matplotlib->pyclesperanto_prototype) (1.4.4) Requirement already satisfied: pytools>=2021.2.7 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyopencl->pyclesperanto_prototype) (2022.1.14) Requirement already satisfied: platformdirs>=2.2.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from pyopencl->pyclesperanto_prototype) (3.2.0) Requirement already satisfied: zipp>=3.1.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from importlib-resources>=3.2.0->matplotlib->pyclesperanto_prototype) (3.15.0) Requirement already satisfied: backcall in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.2.0) Requirement already satisfied: colorama in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.4.6) Requirement already satisfied: typing-extensions in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (4.5.0) Requirement already satisfied: jedi>=0.16 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.18.2) Requirement already satisfied: decorator in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (5.1.1) Requirement already satisfied: stack-data in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.6.2) Requirement already satisfied: pickleshare in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.7.5) Requirement already satisfied: prompt-toolkit!=3.0.37,<3.1.0,>=3.0.30 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (3.0.38) Requirement already satisfied: pygments>=2.4.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (2.15.0) Requirement already satisfied: importlib-metadata>=4.8.3 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from jupyter-client>=6.1.12->ipykernel->stackview) (6.3.0) Requirement already satisfied: pywin32>=300 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from jupyter-core!=5.0.*,>=4.12->ipykernel->stackview) (304) Requirement already satisfied: six>=1.5 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from python-dateutil>=2.7->matplotlib->pyclesperanto_prototype) (1.16.0) Requirement already satisfied: parso<0.9.0,>=0.8.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from jedi>=0.16->ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.8.3) Requirement already satisfied: wcwidth in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from prompt-toolkit!=3.0.37,<3.1.0,>=3.0.30->ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.2.6) Requirement already satisfied: asttokens>=2.1.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (2.2.1) Requirement already satisfied: executing>=1.2.0 in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (1.2.0) Requirement already satisfied: pure-eval in c:\users\rober\appdata\local\mambaforge\envs\bio39\lib\site-packages (from stack-data->ipython>=6.1.0->ipywidgets>=7.6.0->ipycanvas==0.11) (0.2.2)
Again, before going ahead, don't forget to activate the GPU runtime:
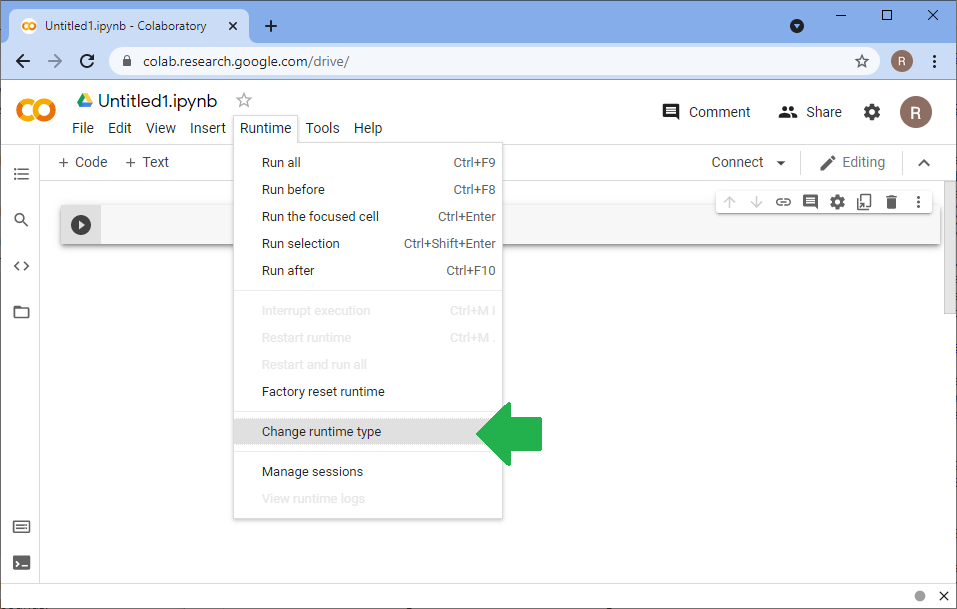
Usage¶
First we test the installation by importing the libraries we're going to use
import stackview
import pyclesperanto_prototype as cle
from skimage.io import imread
# Activate widgets in google colab
from google.colab import output
output.enable_custom_widget_manager()
# check which GPU we are using
cle.select_device("cupy")
# why cupy? https://github.com/clEsperanto/pyclesperanto_prototype/issues/300
<gfx1035 on Platform: AMD Accelerated Parallel Processing (2 refs)>
Starting point is a 3D image dataset provided as numpy array.
image_stack = imread('https://github.com/clEsperanto/clesperanto_example_data/raw/main/Lund_000500_resampled-cropped.tif?raw=true', plugin='tifffile')
image_stack.shape
(100, 256, 256)
cle-images are visualized in jupyer / colab notebooks with a static view. In case the dataset is 3D, we see a maximum-intensity projection.
image_stack_on_gpu = cle.asarray(image_stack)
image_stack_on_gpu
|
|
cle._ image
|
Slicing¶
We can explore the dataset interactively using stackview.slice.
stackview.slice(image_stack_on_gpu)
VBox(children=(HBox(children=(VBox(children=(ImageWidget(height=256, width=256),)),)), IntSlider(value=50, des…
Curtain¶
To compare the original image with a modified version, e.g. after background subtraction, we can use stackview.curtain.
background_subtracted = cle.top_hat_box(image_stack_on_gpu, radius_x=10, radius_y=10)
stackview.curtain(image_stack_on_gpu, background_subtracted)
VBox(children=(HBox(children=(VBox(children=(ImageWidget(height=256, width=256),)),)), IntSlider(value=50, des…
Interaction¶
You can also use sliceview.interact to explore parameters of functions interactively.
stackview.interact(cle.voronoi_otsu_labeling, background_subtracted)
VBox(children=(interactive(children=(FloatSlider(value=2.0, description='spot_sigma', max=25.0, step=1.0), Flo…
This may also make sense with custom functions. When writing those it is important to use type-annotations.
def my_custom_segmentation(image, background_subtraction_radius: int = 10, spot_sigma:float = 1, outline_sigma:float = 1, show_labels: bool = True):
background_subtraction_radius = abs(background_subtraction_radius)
spot_sigma = abs(spot_sigma)
outline_sigma = abs(outline_sigma)
background_subtracted = cle.top_hat_box(image, radius_x=10, radius_y=10)
label_image = cle.voronoi_otsu_labeling(background_subtracted, spot_sigma=spot_sigma, outline_sigma=outline_sigma)
edge_image = cle.detect_label_edges(label_image)
if show_labels:
return label_image
else:
return edge_image * 255 + image
stackview.interact(my_custom_segmentation, image_stack_on_gpu)
VBox(children=(interactive(children=(IntSlider(value=10, description='background_subtraction_radius'), FloatSl…
slice_image = imread('https://github.com/haesleinhuepf/stackview/blob/main/docs/data/blobs.tif?raw=true', plugin='tifffile')
cle.asarray(slice_image)
|
|
cle._ image
|