Cross-Borehole example¶
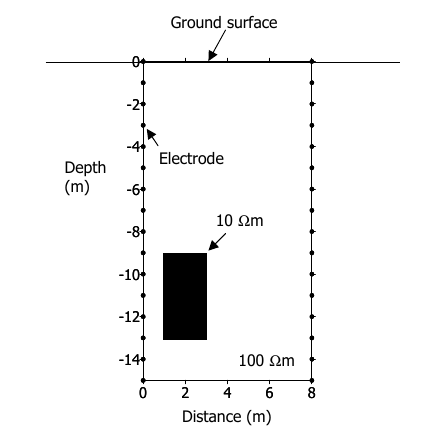
In this example we are going to invert one of the example given in the R2 manual. The aim is to detect a hidden block in the bottom left of the picture as shown below: and this pictures:
In [1]:
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
import os
import sys
import numpy as np # just for parsing the electrode position file
sys.path.append((os.path.relpath('../src'))) # add here the relative path of the API folder
testdir = '../src/examples/dc-2d-borehole/'
from resipy import Project
API path = /media/jkl/data/phd/resipy/src/resipy ResIPy version = 3.4.6 cR2.exe found and up to date. R3t.exe found and up to date. cR3t.exe found and up to date.
Then we will import the protocol.dat file that was outputed by the forward model with this geometry and invert it. Note what we also need to import the electrodes position from a .csv file with 3 columns:x, y, buried. The buried column contains 1 if the electrode is buried and 0 if not.
In [2]:
k = Project(typ='R2')
k.createSurvey(testdir + 'protocol.dat', ftype='ProtocolDC')
k.importElec(testdir + 'elec.csv')
k.createMesh('trian', cl=0.5, cl_factor=20, fmd=20)
# cl is characteristic length, it defines the resolution of the mesh around the electrodes, the smaller, the finer
# cl_factor is how the mesh will grow away from the electrode
# NOTE that a too fine mesh (very small cl) will takes a lot of RAM
# but a too coarse mesh won't be able to resolve the target
k.zlim = [-20, 0]
k.showMesh()
k.invert()
k.showResults(sens=False, contour=True)
Working directory is: /media/jkl/data/phd/resipy/src/resipy
clearing dirname
0/407 reciprocal measurements found.
Creating triangular mesh...done (1114 elements)
Writing .in file and protocol.dat... done
--------------------- MAIN INVERSION ------------------
>> R 2 R e s i s t i v i t y I n v e r s i o n v4.10 <<
>> D a t e : 03 - 12 - 2023
>> My beautiful survey
>> I n v e r s e S o l u t i o n S e l e c t e d <<
>> Determining storage needed for finite element conductance matrix
>> Generating index array for finite element conductance matrix
>> Reading start resistivity from res0.dat
>> R e g u l a r i s e d T y p e <<
>> L i n e a r F i l t e r <<
>> L o g - D a t a I n v e r s i o n <<
>> N o r m a l R e g u l a r i s a t i o n <<
>> D a t a w e i g h t s w i l l b e m o d i f i e d <<
Processing dataset 1
Measurements read: 407 Measurements rejected: 0
Geometric mean of apparent resistivities: 0.92723E+02
>> Total Memory required is: 0.004 Gb
Iteration 1
Initial RMS Misfit: 6.25 Number of data ignored: 1
Alpha: 1404.342 RMS Misfit: 2.49 Roughness: 0.773
Alpha: 651.838 RMS Misfit: 2.14 Roughness: 1.435
Alpha: 302.556 RMS Misfit: 1.82 Roughness: 2.544
Alpha: 140.434 RMS Misfit: 1.58 Roughness: 4.135
Alpha: 65.184 RMS Misfit: 1.41 Roughness: 6.322
Alpha: 30.256 RMS Misfit: 1.28 Roughness: 9.578
Alpha: 14.043 RMS Misfit: 1.19 Roughness: 14.884
Alpha: 6.518 RMS Misfit: 1.19 Roughness: 23.529
Step length set to 1.00000
Final RMS Misfit: 1.19
Attempted to update data weights and caused overshoot
treating as converged
Solution converged - Outputing results to file
Calculating sensitivity map
Processing dataset 2
End of data: Terminating
1/1 results parsed (1 ok; 0 failed)
All ok /media/jkl/data/phd/resipy/doc/gallery/../../src/resipy/meshTools.py:1487: MatplotlibDeprecationWarning: The collections attribute was deprecated in Matplotlib 3.8 and will be removed two minor releases later. for col in cont.collections: /media/jkl/data/phd/resipy/doc/gallery/../../src/resipy/Project.py:4136: MatplotlibDeprecationWarning: The collections attribute was deprecated in Matplotlib 3.8 and will be removed two minor releases later. colls = mesh.cax.collections if contour == True else [mesh.cax]
In [ ]: