How Jupyter works¶
...But first, Terminal IPython¶
When you type ipython, you get the original IPython interface, running in
the terminal. It does something like this:
while True:
code = input(">>> ")
exec(code)
Of course, it's much more complex, because it has to deal with multi-line
code, tab completion using readline or prompt-toolkit, magic commands, and so on. But the
model is like that: prompt the user for some code, and when they've entered it,
exec it in the same process. This model is often called a REPL, or
Read-Eval-Print-Loop.
Jupyter and The IPython Kernel¶
Jupyter expands on this REPL model and provides other interfaces to running code–the notebook, the Qt console, jupyter console in
the terminal, and third party interfaces—use the IPython Kernel. This is a
separate process which is responsible for running user code, and things like
computing possible completions. Frontends communicate with it using JSON
messages sent over ZeroMQ sockets; the protocol they use is described in
the jupyter protocol.
The core execution machinery for the kernel is shared with terminal IPython:
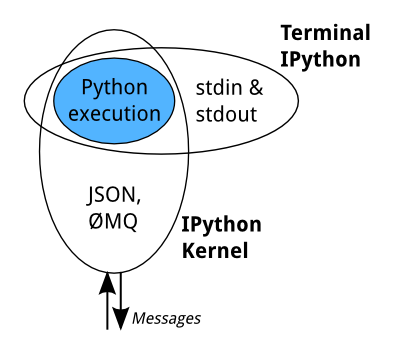
A kernel process can be connected to more than one frontend simultaneously. In this case, the different frontends will have access to the same variables.
This design was intended to allow easy development of different frontends based on the same kernel, but it also made it possible to support new languages in the same frontends, by developing kernels in those languages, and we are refining IPython to make that more practical.
Today, there are two ways to develop a kernel for another language. Wrapper kernels reuse the communications machinery from IPython, and implement only the core execution part. Native kernels implement execution and communications in the target language:
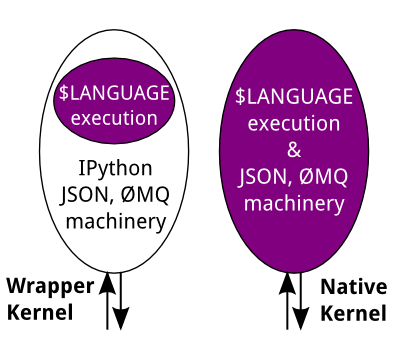
Wrapper kernels are easier to write quickly for languages that have good Python wrappers, like octave_kernel, or languages where it's impractical to implement the communications machinery, like bash_kernel. Native kernels are likely to be better maintained by the community using them, like IJulia or IHaskell.
Notebooks¶
The Notebook frontend does something extra. In addition to running your code, it
stores code and output, together with markdown notes, in an editable document
called a notebook. When you save it, this is sent from your browser to the
notebook server, which saves it on disk as a JSON file with a .ipynb
extension.
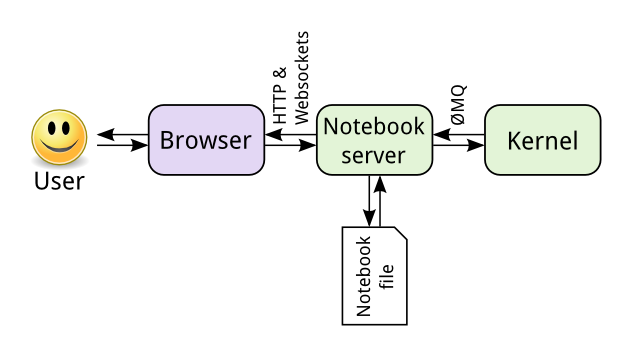
The notebook server, not the kernel, is responsible for saving and loading notebooks, so you can edit notebooks even if you don't have the kernel for that language—you just won't be able to run code. The kernel doesn't know anything about the notebook document: it just gets sent cells of code to execute when the user runs them.
Install more languages¶
The exact procedure to install a kernel for a different language will depend on the specificity of each language. Though ther is a common set of step to follow.
- Install the language stack you are interested in.
- Install the kernel for this language (often using given language package manager).
- Register the kernel globally with Jupyter.
While usually a kernel is though as a specific language, a kernel may be:
- A virtual environment (or equivalent)
- A set of configuration/environment variables.
- A physical location (for remote kernels)
Installing multiple kernels does not automatically allow one notebook to use many languages at once, but this is also possible.
A community maintained list of available kernel can be found on the Jupyter Wiki.
Install a second python kernel¶
Using conda:
$ conda create -n pycon-env python=3.6 --yes --quiet
$ source activate pycon-env
(pycon-env)$ conda install ipykernel --yes
(pycon-env)$ python -m ipykernel install --name pycon-kernel
Installed kernelspec pycon-kernel in /usr/local/share/jupyter/kernels/pycon-kernel
Available options are --user, --name <machine-readable-name>, --display-name <"User Friendly Name">
Install and R kernel¶
Again using conda, let's install the R stack and create an R kernel.
In a shell:
$ conda install -c r r # install r form teh R channel
$ conda install -c r r-irkernel
$ r
> IRkernel::installspec()
If you are not using conda you may need to replace by :
$ R
> install.packages(c('repr', 'IRdisplay', 'evaluate', 'crayon', 'pbdZMQ', 'devtools', 'uuid', 'digest'))
...
> devtools::install_github('IRkernel/IRkernel')
You may want to install Rin the same environment as the previous Python Kernel if you wish to do R and python inthe same notebook.
install more kernels¶
Feel free to experiment with other kernel, poke at the installed kernelspec folders. Use the following to list all the kernels and their locations:
$ jupyter kernelspec list
Available kernels:
ir /home/jovyan/.local/share/jupyter/kernels/ir
julia-0.5 /opt/conda/share/jupyter/kernels/julia-0.5
python3 /opt/conda/share/jupyter/kernels/python3
python2 /usr/local/share/jupyter/kernels/python2
The above process can take a long time, need to compile a few modules. You may want to try the jupyter/datascience-notebook Docker image which already have Python, Julia and R installed. Warning the Docker image is big (several GB) ! Please don't try to download it on Conference wifi.