Elements of Evolutionary Algorithms¶
The notebook is better viewed rendered as slides. You can convert it to slides and view them by:
- using nbconvert with a command like:
$ ipython nbconvert --to slides --post serve <this-notebook-name.ipynb>
- installing Reveal.js - Jupyter/IPython Slideshow Extension
- using the online IPython notebook slide viewer (some slides of the notebook might not be properly rendered).
This and other related IPython notebooks can be found at the course github repository:
Elements to take into account using evolutionary algorithms¶
- Individual representation (binary, Gray, floating-point, etc.);
- evaluation and fitness assignment;
- mating selection, that establishes a partial order of individuals in the population using their fitness function value as reference and determines the degree at which individuals in the population will take part in the generation of new (offspring) individuals.
- variation, that applies a range of evolution-inspired operators, like crossover, mutation, etc., to synthesize offspring individuals from the current (parent) population. This process is supposed to prime the fittest individuals so they play a bigger role in the generation of the offspring.
- environmental selection, that merges the parent and offspring individuals to produce the population that will be used in the next iteration. This process often involves the deletion of some individuals using a given criterion in order to keep the amount of individuals bellow a certain threshold.
- stopping criterion, that determines when the algorithm shoulod be stopped, either because the optimum was reach or because the optimization process is not progressing.
Hence a 'general' evolutionary algorithm can be described as¶
def evolutionary_algorithm():
'Pseudocode of an evolutionary algorithm'
populations = [] # a list with all the populations
populations[0] = initialize_population(pop_size)
t = 0
while not stop_criterion(populations[t]):
fitnesses = evaluate(populations[t])
offspring = matting_and_variation(populations[t],
fitnesses)
populations[t+1] = environmental_selection(
populations[t],
offspring)
t = t+1
Python libraries for evolutionary computation¶
- PaGMO/PyGMO
- Inspyred
- Distributed Evolutionary Algorithms in Python (DEAP)
There are potentially many more, feel free to give me some feedback on this.

|
|
Lets start with an example and analyze it¶
The One Max problem¶
- Maximize the number of ones in a binary string (list, vector, etc.).
- More formally, from the set of binary strings of length $n$,
- Find $s^\ast\in\mathcal{S}$ such that
- Its clear that the optimum is an all-ones string.
Coding the problem¶
import random
from deap import algorithms, base, creator, tools
import matplotlib.pyplot as plt
%matplotlib inline
%config InlineBackend.figure_format = 'retina'
plt.rc('text', usetex=True)
plt.rc('font', family='serif')
plt.rcParams['text.latex.preamble'] ='\\usepackage{libertine}\n\\usepackage[utf8]{inputenc}'
import seaborn
seaborn.set(style='whitegrid')
seaborn.set_context('notebook')
creator.create("FitnessMax", base.Fitness, weights=(1.0,))
creator.create("Individual", list, fitness=creator.FitnessMax)
def evalOneMax(individual):
return (sum(individual),)
Defining the elements¶
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual,
toolbox.attr_bool, n=100)
toolbox.register("population", tools.initRepeat, list,
toolbox.individual)
toolbox.register("evaluate", evalOneMax)
toolbox.register("mate", tools.cxTwoPoint)
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
toolbox.register("select", tools.selTournament, tournsize=3)
Running the experiment¶
pop = toolbox.population(n=300)
Lets run only 10 generations
result = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2,
ngen=10, verbose=False)
print('Current best fitness:', evalOneMax(tools.selBest(pop, k=1)[0]))
Current best fitness: (83,)
result = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2,
ngen=50, verbose=False)
print('Current best fitness:', evalOneMax(tools.selBest(pop, k=1)[0]))
Current best fitness: (100,)
Essential features¶
deap.creator: meta-factory allowing to create classes that will fulfill the needs of your evolutionary algorithms.deap.base.Toolbox: A toolbox for evolution that contains the evolutionary operators. You may populate the toolbox with any other function by using theregister()methoddeap.base.Fitness([values]): The fitness is a measure of quality of a solution. If values are provided as a tuple, the fitness is initalized using those values, otherwise it is empty (or invalid). You should inherit from this class to define your custom fitnesses.
Defining an individual¶
First import the required modules and register the different functions required to create individuals that are a list of floats with a minimizing two objectives fitness.
import random
from deap import base
from deap import creator
from deap import tools
IND_SIZE = 5
creator.create("FitnessMin", base.Fitness, weights=(-1.0, -1.0))
creator.create("Individual", list, fitness=creator.FitnessMin)
toolbox1 = base.Toolbox()
toolbox1.register("attr_float", random.random)
toolbox1.register("individual", tools.initRepeat, creator.Individual,
toolbox1.attr_float, n=IND_SIZE)
/Users/lm/anaconda/lib/python3.6/site-packages/deap-1.1.0-py3.6-macosx-10.7-x86_64.egg/deap/creator.py:141: RuntimeWarning: A class named 'Individual' has already been created and it will be overwritten. Consider deleting previous creation of that class or rename it. RuntimeWarning)
The first individual can now be built
ind1 = toolbox1.individual()
Printing the individual ind1 and checking if its fitness is valid will give something like this
print(ind1)
[0.2989651981634037, 0.9689899788264542, 0.3524509946515799, 0.06381306368762452, 0.4555534543646952]
print(ind1.fitness.valid)
False
The individual is printed as its base class representation (here a list) and the fitness is invalid because it contains no values.
Evaluation¶
The evaluation is the most "personal" part of an evolutionary algorithm
- it is the only part of the library that you must write yourself.
- A typical evaluation function takes one individual as argument and return its fitness as a tuple.
- A fitness is a list of floating point values and has a property valid to know if this individual shall be re-evaluated.
- The fitness is set by setting the values to the associated tuple.
For example, the following evaluates the previously created individual ind1 and assign its fitness to the corresponding values.
def evaluate(individual):
# Do some hard computing on the individual
a = sum(individual)
b = len(individual)
return a, 1. / b
ind1.fitness.values = evaluate(ind1)
print(ind1.fitness.valid)
True
print(ind1.fitness)
(2.139772689693758, 0.2)
Dealing with single objective fitness is not different, the evaluation function must return a tuple because single-objective is treated as a special case of multi-objective.
Mutation¶
- The next kind of operator that we will present is the mutation operator.
- There is a variety of mutation operators in the deap.tools module.
- Each mutation has its own characteristics and may be applied to different type of individual.
- Be careful to read the documentation of the selected operator in order to avoid undesirable behaviour.
The general rule for mutation operators is that they only mutate, this means that an independent copy must be made prior to mutating the individual if the original individual has to be kept or is a reference to an other individual (see the selection operator).
In order to apply a mutation (here a gaussian mutation) on the individual ind1, simply apply the desired function.
mutant = toolbox1.clone(ind1)
ind2, = tools.mutGaussian(mutant, mu=0.0, sigma=0.2, indpb=0.2)
del mutant.fitness.values
The fitness’ values are deleted because they not related to the individual anymore. As stated above, the mutation does mutate and only mutate an individual it is not responsible of invalidating the fitness nor anything else. The following shows that ind2 and mutant are in fact the same individual.
ind2 is mutant
True
mutant is ind2
True
Crossover¶
- There is a variety of crossover operators in the
deap.tools module. - Each crossover has its own characteristics and may be applied to different type of individuals.
- Be careful to read the documentation of the selected operator in order to avoid undesirable behaviour.
The general rule for crossover operators is that they only mate individuals, this means that an independent copies must be made prior to mating the individuals if the original individuals have to be kept or is are references to other individuals (see the selection operator).
Lets apply a crossover operation to produce the two children that are cloned beforehand.
child1, child2 = [toolbox1.clone(ind) for ind in (ind1, ind2)]
tools.cxBlend(child1, child2, 0.5)
del child1.fitness.values
del child2.fitness.values
Selection¶
- Selection is made among a population by the selection operators that are available in the deap.operators module.
- The selection operator usually takes as first argument an iterable container of individuals and the number of individuals to select. It returns a list containing the references to the selected individuals.
The selection is made as follow.
selected = tools.selBest([child1, child2], 2)
child1 in selected
True
Using the Toolbox¶
- The toolbox is intended to contain all the evolutionary tools, from the object initializers to the evaluation operator.
- It allows easy configuration of each algorithms.
- The toolbox has basically two methods,
register()andunregister(), that are used to add or remove tools from the toolbox. - The usual names for the evolutionary tools are mate(), mutate(), evaluate() and select(), however, any name can be registered as long as it is unique. Here is how they are registered in the toolbox.
from deap import base
from deap import tools
toolbox1 = base.Toolbox()
def evaluateInd(individual):
# Do some computation
return result,
toolbox1.register("mate", tools.cxTwoPoint)
toolbox1.register("mutate", tools.mutGaussian, mu=0, sigma=1, indpb=0.2)
toolbox1.register("select", tools.selTournament, tournsize=3)
toolbox1.register("evaluate", evaluateInd)
Tool Decoration¶
- A powerful feature that helps to control very precise thing during an evolution without changing anything in the algorithm or operators.
- A decorator is a wrapper that is called instead of a function.
- It is asked to make some initialization and termination work before and after the actual function is called.
For example, in the case of a constrained domain, one can apply a decorator to the mutation and crossover in order to keep any individual from being out-of-bound.
The following defines a decorator that checks if any attribute in the list is out-of-bound and clips it if it is the case.
- The decorator is defined using three functions in order to receive the min and max arguments.
- Whenever the mutation or crossover is called, bounds will be check on the resulting individuals.
def checkBounds(min, max):
def decorator(func):
def wrapper(*args, **kargs):
offspring = func(*args, **kargs)
for child in offspring:
for i in xrange(len(child)):
if child[i] > max:
child[i] = max
elif child[i] < min:
child[i] = min
return offspring
return wrapper
return decorator
toolbox.register("mate_example", tools.cxBlend, alpha=0.2)
toolbox.register("mutate_example", tools.mutGaussian, mu=0, sigma=2)
MIN = 0; MAX = 10
toolbox.decorate("mate_example", checkBounds(MIN, MAX))
toolbox.decorate("mutate_example", checkBounds(MIN, MAX))
This will work on crossover and mutation because both return a tuple of individuals. The mutation is often considered to return a single individual but again like for the evaluation, the single individual case is a special case of the multiple individual case.
Variations¶
- Variations allows to build simple algorithms using predefined small building blocks.
- In order to use a variation, the toolbox must be set to contain the required operators.
For example, in the lastly presented complete algorithm, the crossover and mutation are regrouped in the
varAnd()function, this function requires the toolbox to contain themate()andmutate()functions. The variations can be used to simplify the writing of an algorithm as follow.
from deap import algorithms
NGEN = 20 # number of generations
CXPB = 0.6
MUTPB = 0.05
for g in range(NGEN):
# Select and clone the next generation individuals
offspring = map(toolbox.clone, toolbox.select(pop, len(pop)))
# Apply crossover and mutation on the offspring
offspring = algorithms.varAnd(offspring, toolbox, CXPB, MUTPB)
# Evaluate the individuals with an invalid fitness
invalid_ind = [ind for ind in offspring if not ind.fitness.valid]
fitnesses = toolbox.map(toolbox.evaluate, invalid_ind)
for ind, fit in zip(invalid_ind, fitnesses):
ind.fitness.values = fit
# The population is entirely replaced by the offspring
pop[:] = offspring
Algorithms¶
- There are several algorithms implemented in the algorithms module.
- They are very simple and reflect the basic types of evolutionary algorithms present in the literature.
- The algorithms use a Toolbox as defined in the last sections.
- In order to setup a toolbox for an algorithm, you must register the desired operators under a specified names, refer to the documentation of the selected algorithm for more details.
- Once the toolbox is ready, it is time to launch the algorithm.
The simple evolutionary algorithm takes 5 arguments, a population, a toolbox, a probability of mating each individual at each generation (cxpb), a probability of mutating each individual at each generation (mutpb) and a number of generations to accomplish (ngen).
from deap import algorithms
result = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=50)
gen nevals 0 0 1 176 2 171 3 177 4 188 5 197 6 170 7 175 8 178 9 178 10 190 11 183 12 169 13 189 14 187 15 161 16 175 17 155 18 179 19 171 20 182 21 187 22 175 23 170 24 207 25 173 26 176 27 196 28 216 29 167 30 173 31 182 32 167 33 188 34 179 35 203 36 162 37 174 38 191 39 179 40 183 41 190 42 187 43 204 44 193 45 177 46 191 47 170 48 161 49 158 50 194
Computing Statistics¶
Often, one wants to compile statistics on what is going on in the optimization. The Statistics are able to compile such data on arbitrary attributes of any designated object. To do that, one need to register the desired statistic functions inside the stats object using the exact same syntax as the toolbox.
stats = tools.Statistics(key=lambda ind: ind.fitness.values)
The statistics object is created using a key as first argument. This key must be supplied a function that will later be applied to the data on which the statistics are computed. The previous code sample uses the fitness.values attribute of each element.
import numpy
stats.register("avg", numpy.mean)
stats.register("std", numpy.std)
stats.register("min", numpy.min)
stats.register("max", numpy.max)
- The statistical functions are now registered.
- The register function expects an alias as first argument and a function operating on vectors as second argument.
- Any subsequent argument is passed to the function when called. The creation of the statistics object is now complete.
Predefined Algorithms¶
When using a predefined algorithm such as eaSimple(), eaMuPlusLambda(), eaMuCommaLambda(), or eaGenerateUpdate(), the statistics object previously created can be given as argument to the algorithm.
pop, logbook = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=0,
stats=stats, verbose=True)
gen nevals avg std min max 0 0 98.8 2.47521 88 100
- Statistics will automatically be computed on the population every generation.
- The verbose argument prints the statistics on screen while the optimization takes place.
- Once the algorithm returns, the final population and a Logbook are returned.
- See the next section or the Logbook documentation for more information.
Writing Your Own Algorithm¶
When writing your own algorithm, including statistics is very simple. One need only to compile the statistics on the desired object.
For example, compiling the statistics on a given population is done by calling the compile() method.
record = stats.compile(pop)
The argument to the compile function must be an iterable of elements on which the key will be called. Here, our population (pop) contains individuals.
- The statistics object will call the key function on every individual to retrieve their
fitness.valuesattribute. - The resulting array of values is finally given the each statistic function and the result is put into the record dictionary under the key associated with the function.
- Printing the record reveals its nature.
print(record)
{'avg': 98.799999999999997, 'std': 2.4752104287649295, 'min': 88.0, 'max': 100.0}
Logging Data¶
Once the data is produced by the statistics, one can save it for further use in a Logbook.
- The logbook is intended to be a chronological sequence of entries (as dictionaries).
- It is directly compliant with the type of data returned by the statistics objects, but not limited to this data.
- In fact, anything can be incorporated in an entry of the logbook.
logbook = tools.Logbook()
logbook.record(gen=0, evals=30, **record)
The record() method takes a variable number of argument, each of which is a data to be recorded. In the last example, we saved the generation, the number of evaluations and everything contained in the record produced by a statistics object using the star magic. All record will be kept in the logbook until its destruction.
After a number of records, one may want to retrieve the information contained in the logbook.
gen, avg = logbook.select("gen", "avg")
The select() method provides a way to retrieve all the information associated with a keyword in all records. This method takes a variable number of string arguments, which are the keywords used in the record or statistics object. Here, we retrieved the generation and the average fitness using a single call to select.
Printing to Screen¶
- A logbook can be printed to screen or file.
- Its
__str__()method returns a header of each key inserted in the first record and the complete logbook for each of these keys. - The row are in chronological order of insertion while the columns are in an undefined order.
- The easiest way to specify an order is to set the header attribute to a list of strings specifying the order of the columns.
logbook.header = "gen", "avg", "spam"
The result is:
print(logbook)
gen avg spam 0 98.8
Plotting Features¶
- One of the most common operation when an optimization is finished is to plot the data during the evolution.
- The
Logbookallows to do this very efficiently. - Using the select method, one can retrieve the desired data and plot it using matplotlib.
pop = toolbox.population(n=300)
pop, logbook = algorithms.eaSimple(pop, toolbox, cxpb=0.5, mutpb=0.2, ngen=50,
stats=stats, verbose=False)
gen = logbook.select("gen")
fit_mins = logbook.select("min")
size_avgs = logbook.select("avg")
fig, ax1 = plt.subplots()
line1 = ax1.plot(gen, fit_mins, "b-", label="Minimum Fitness")
ax1.set_xlabel("Generation")
ax1.set_ylabel("Fitness", color="b")
for tl in ax1.get_yticklabels():
tl.set_color("b")
ax2 = ax1.twinx()
line2 = ax2.plot(gen, size_avgs, "r-", label="Average Fitness")
ax2.set_ylabel("Size", color="r")
for tl in ax2.get_yticklabels():
tl.set_color("r")
lns = line1 + line2
labs = [l.get_label() for l in lns]
ax1.legend(lns, labs, loc="lower right", frameon=True)
plt.show()
Constraint Handling¶
We have already seen some alternatives.
- Penality functions are the most basic way of handling constrains for individuals that cannot be evaluated or are forbiden for problem specific reasons, when falling in a given region.
- The penality function gives a fitness disavantage to theses individuals based on the amount of constraint violation in the solution.
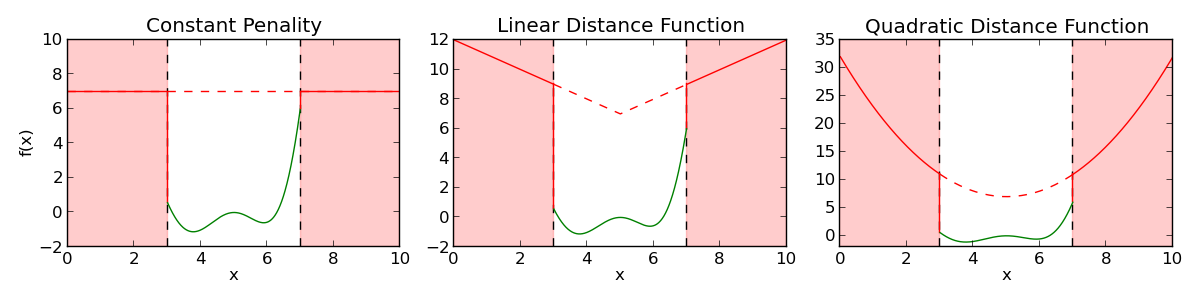
In DEAP, a penality function can be added to any evaluation function using the DeltaPenality decorator provided in the tools module.
from math import sin
from deap import base
from deap import tools
def evalFct(individual):
"""Evaluation function for the individual."""
x = individual[0]
return (x - 5)**2 * sin(x) * (x/3),
def feasible(individual):
"""Feasability function for the individual. Returns True if feasible False
otherwise."""
if 3 < individual[0] < 5:
return True
return False
def distance(individual):
"""A distance function to the feasability region."""
return (individual[0] - 5.0)**2
toolbox = base.Toolbox()
toolbox.register("evaluate", evalFct)
toolbox.decorate("evaluate", tools.DeltaPenality(feasible, 7.0, distance))

# To install run: pip install version_information
%load_ext version_information
%version_information scipy, numpy, matplotlib, seaborn, deap
| Software | Version |
|---|---|
| Python | 3.6.0 64bit [GCC 4.2.1 Compatible Apple LLVM 6.0 (clang-600.0.57)] |
| IPython | 5.2.2 |
| OS | Darwin 16.4.0 x86_64 i386 64bit |
| scipy | 0.18.1 |
| numpy | 1.11.3 |
| matplotlib | 2.0.0 |
| seaborn | 0.7.1 |
| deap | 1.1 |
| Sat Mar 04 03:52:24 2017 BRT | |
# this code is here for cosmetic reasons
from IPython.core.display import HTML
from urllib.request import urlopen
HTML(urlopen('https://raw.githubusercontent.com/lmarti/jupyter_custom/master/custom.include').read().decode('utf-8'))

