Groupby Architecture¶
Problem¶
Pandas has a well-designed groupby architecture, but when developing against it I often hit three challenges:
- It involves 4 to 5 classes, which can be hard to keep track of.
- Its design is similar to Categoricals--but what class names
codes, another might namelabels. - Correctly ordering results uses a somewhat uncommon class: Splitter.
This document lays out...
- The big picture behind the groupby architecture.
- 3 key splitting cases: multiple group columns, NA group keys, and regrouping.
- How to order results for combining aggregation and transformation.
By the end of this document, a person should be able to construct fast, custom split-apply-combine operations that perform over numpy arrays.
Motivating scenario¶
Consider the corr() method. This makes it easy to correlate two pandas Series.
However, corr() is not available for grouped Series.
from siuba.data import mtcars
g_cyl = mtcars.groupby('cyl')
# works
mtcars.hp.corr(mtcars.mpg)
# doesn't work
# g_cyl.hp.corr(g_cyl.mpg)
-0.7761683718265864
The corr() method largely just punts the operation to numpy, so in theory it shouldn't be hard to implement over grouped data. However, no method involving two grouped Series is implemented, which is why this doc exists.
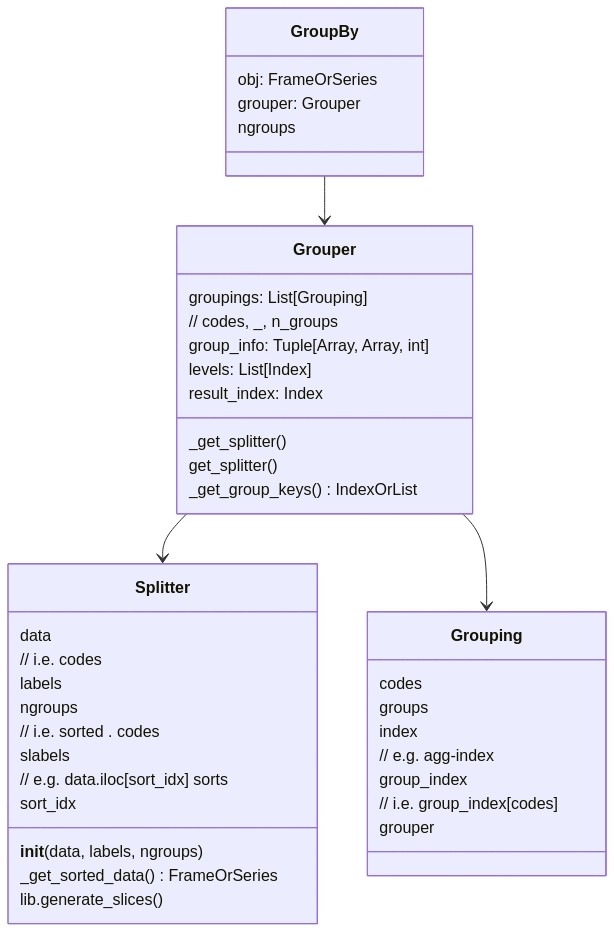
Big picture¶
Below is a class diagrom for the classes involved.
This diagram is a sketch, to help with the sequence diagram focused on splitting in the next section, so includes most, but not all properties. Lines starting with // are comments.
Class Responsibility-Collaboration Cards (wiki):
- Grouper
- collect all information for group splits (e.g. over multiple columns)
- serve as primary point of interaction (e.g.
get_splitter(),result_index)
- Grouping
- Represent an individual grouping column / its splitting info
- Represent index for individual aggregation result index
- Splitter
- Calculate arrays for sorting and unsorting operations
- Sort data according to group keys
- Calculate group slices for sorted data
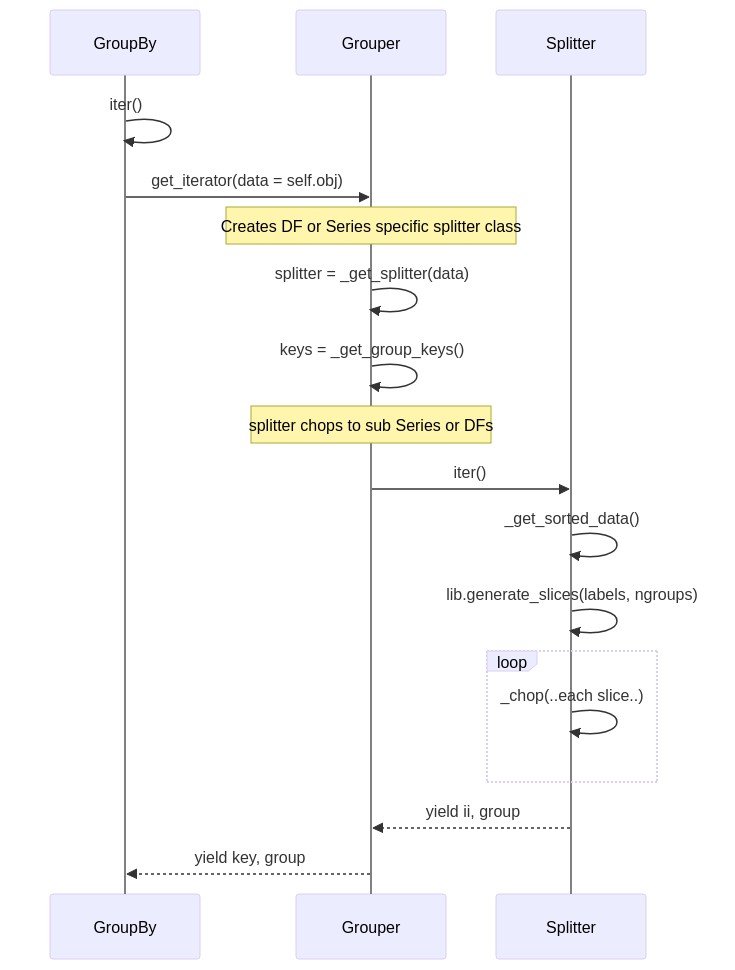
Splitting¶
The sequence diagram below gives a flavor for how splitting occurs, based on the scenario where you loop over grouped data.
Below is a complete example of running the split manually, but by chopping numpy arrays directly.
import pandas as pd
from pandas._libs import lib
df = pd.DataFrame({
'g': ['c', 'b', 'b', 'a'],
'x': [1,2,3,4]
}, index = [10, 11, 12, 13])
gdf = df.groupby('g')
# note grouped Series and DataFrame have the same Grouper
splitter = gdf.x.grouper._get_splitter(gdf.x.obj)
starts, ends = lib.generate_slices(splitter.slabels, splitter.ngroups)
arr_x = splitter._get_sorted_data().values
# iterate over splits, applying function
keys = gdf.grouper._get_group_keys()
results = []
for i, (start, end) in enumerate(zip(starts, ends)):
print("Group:", i, keys[i])
print(arr_x[start:end])
Group: 0 a [4] Group: 1 b [2 3] Group: 2 c [1]
Grouping by multiple columns¶
TODO
NAs in grouping columns¶
Note that by default pandas groupby drops rows where a grouping column has NAs.
import pandas as pd
df = pd.DataFrame({'g': ['a', 'a', pd.NA], 'x': [1,2,3]})
# Only has result for 'a' group
df.groupby('g').agg('mean')
| x | |
|---|---|
| g | |
| a | 1.5 |
# Keeps groups w/ NAs
df.groupby('g', dropna = False).agg('mean')
| x | |
|---|---|
| g | |
| a | 1.5 |
| NaN | 3.0 |
TODO: How does this show up in the groupby classes?
Regrouping¶
One really useful property is that you can pass a Grouper to the groupby method.
g_cyl2 = g_cyl.obj.groupby(g_cyl.grouper)
g_cyl.grouper is g_cyl2.grouper
True
This is how siuba is able to regroup transformations and compose operations like below.
from siuba.experimental.pd_groups import fast_mutate
from siuba import _
## doesn't work
#(g_cyl.mpg + g_cyl.mpg) / g_cyl.mpg
# fine in siuba, regroups after each operation
fast_mutate(g_cyl, res = (_.mpg + _.mpg) / _.hp)
# essentially runs
op_res1 = (g_cyl.obj.mpg + g_cyl.obj.mpg).groupby(g_cyl.grouper)
op_res2 = op_res1.obj / g_cyl.obj.hp
op_res2.head()
0 0.381818 1 0.381818 2 0.490323 3 0.389091 4 0.213714 dtype: float64
Applying¶
In general, pandas apply architecture is very complicated. Much of the strategy involves...
- looking up ideal forms of an operation from a string. e.g. .agg('mean').
- trying things the fast way first, to see if they work.
Because there is an incredible amount to be gained by applying operations to only numpy arrays, we won't go into more details here on how pandas does applies. Keep in mind that splitting and performing operations on numpy arrays tends to be very fast.
Combine variants¶
The following sections show how the groupby architecture can aggregate different kinds of results. I'll use results that are numpy arrays for these examples.
Key points for creating new columns from grouped operations:
- a manual split (e.g. via a loop over GroupBy) sorts chunks by group levels.
- an agg uses the grouping columns as its index, and sorts by them.
- take Grouper.codes (or Splitter.labels) to convert aggregation to original order.
- take Splitter.sort_idx to convert full length result over groups to original order.
import pandas as pd
df = pd.DataFrame({
"g": ['b', 'c', 'a', 'c'],
"x": [10, 11, 12, 13]
}, index = [100, 101, 102, 103]
)
gdf = df.groupby('g')
Aggregate¶
The pandas agg method returns a Series with 1 result per group. The results are sorted by their index, which is the grouping columns.
res_agg = gdf.x.agg('mean')
res_agg
g a 12 b 10 c 12 Name: x, dtype: int64
- result is sorted by the index (e.g. df.sort_index())
- the grouper property
result_indexis the index for aggregations
# result index and grouper property are the same object
res_agg.index is gdf.grouper.result_index
True
# recreating Series from numpy array (values)
pd.Series(res_agg.values, gdf.grouper.result_index)
g a 12 b 10 c 12 dtype: int64
# recreating same aggregation using a loop
import numpy as np
res_arr = np.array([g['x'].values.mean() for k, g in gdf])
res_arr
array([12., 10., 12.])
pd.Series(res_arr, gdf.grouper.result_index)
g a 12.0 b 10.0 c 12.0 dtype: float64
Transform (agg op)¶
The pandas transform method is like a dplyr mutate. Its final result is returned in the same order as the original data.
res_trans1 = gdf.x.transform('mean')
res_trans1
100 10 101 12 102 12 103 12 Name: x, dtype: int64
res_agg
g a 12 b 10 c 12 Name: x, dtype: int64
codes, _, ngroups = gdf.grouper.group_info
# which category does each element belong to?
codes
array([1, 2, 0, 2])
# note pandas uses pandas.core.algorithms.take_1d
# could also use res_agg.iloc[codes] or Splitter.labels (shown below)
res_agg.take(codes)
res_agg.values[codes]
array([10, 12, 12, 12])
Note that a manual transformed aggregate can be done as follows.
res = gdf.x.mean()
# grouper.group_info is a 3-tuple: (ids, _, n_groups)
res.take(gdf.grouper.group_info[0])
g b 10 c 12 a 12 c 12 Name: x, dtype: int64
Transform (window op)¶
Elementwise operations, like x + x aren't possible with groupby objects, so we'll consider the cumulative sum operation, cumsum. Unlike mean, it calculates one result per element in the series.
res_trans2 = gdf.x.transform('cumsum')
res_trans2
100 10 101 11 102 12 103 24 Name: x, dtype: int64
res_trans2.values
array([10, 11, 12, 24])
res_arr = np.concatenate([x.cumsum().values for k, x in gdf.x]).ravel()
res_arr
array([12, 10, 11, 24])
Notice that the result above is sorted by group, with the value for the first group ("a") first.
In order to get back the original order, like in transform, we use splitter.sort_idx.argsort().
splitter = gdf.grouper._get_splitter(gdf.obj)
res_arr.take(splitter.sort_idx.argsort())
array([10, 11, 12, 24])
Essentially, sort_idx was the order used to sort the original result, and argsort reverses it!
# taking this moves
# * position 2 to position 0
splitter.sort_idx
array([2, 0, 1, 3])
# taking this moves the position 0 to position 2
splitter.sort_idx.argsort()
array([1, 2, 0, 3])
Filtering (the dplyr version)¶
As far as I know, there is now simple mechanism for filtering grouped data (other than running groupby again).
Summary¶
- aggs have
grouper.result_indexset as their index. - aggs: get transformed, original order using
grouper.group_info[0]. - windows: get original order using
splitter.sort_idx.argsort().