Accessing Forest Inventory and Analysis data with the Planetary Computer STAC API¶
This notebook demonstrates accessing Forest Inventory and Analysis (FIA) data from the Planetary Computer STAC API.
The Forest Inventory and Analysis collection contains many tables, and each STAC table corresponds to one STAC item in the FIA collection. In this example, we'll use a few of the tables to estimate the total amount of aboveground carbon, in pounds, per US county.
This example builds on the plot estimation example from the rfia package.
from cartopy import crs as ccrs
import dask_gateway
import dask_geopandas
import dask.dataframe as dd
import geopandas
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import pystac_client
import planetary_computer
The tree table below is relatively large, so we'll process it in parallel on a Dask cluster. The example will still run without a cluster, it will just take longer.
cluster = dask_gateway.GatewayCluster()
cluster.scale(16)
client = cluster.get_client()
cluster
VBox(children=(HTML(value='<h2>GatewayCluster</h2>'), HBox(children=(HTML(value='\n<div>\n<style scoped>\n …
Data Access¶
We'll use two datsaets
tree: Information on each tree 1″ in diameter or larger, linked toplotandcond.plot: Information relevant to each 1-acre field plot where the samples were collected.
All of these are available in Azure Blob Storage as parquet datasets that can be read, for example, by dask.dataframe.
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
)
fia = catalog.get_collection("fia")
fia
<CollectionClient id=fia>
plot_item = fia.get_item("plot")
tree_item = fia.get_item("tree")
And now we can read the items using dask.dataframe. We make sure to pass the storage_options included in the data asset.
plot = dd.read_parquet(
plot_item.assets["data"].href,
columns=["CN", "STATECD", "COUNTYCD"],
storage_options=plot_item.assets["data"].extra_fields["table:storage_options"],
engine="pyarrow",
)
tree = dd.read_parquet(
tree_item.assets["data"].href,
columns=["PLT_CN", "CONDID", "TREE", "DRYBIO_AG", "CARBON_AG", "TPA_UNADJ"],
storage_options=tree_item.assets["data"].extra_fields["table:storage_options"],
engine="pyarrow",
filters=[("CONDID", "==", 1)],
)
Join the datasets¶
The three datasets can be joined on their various keys. Since tree is relatively large, we'll join the other (smaller, in-memory) dataframes to it.
df = tree.merge(plot.assign(PLT_CN=plot.CN).compute(), on="PLT_CN").assign(
bio=lambda df: df.DRYBIO_AG * df.TPA_UNADJ / 2000,
carbon=lambda df: df.CARBON_AG * df.TPA_UNADJ / 2000,
)
df
| PLT_CN | CONDID | TREE | DRYBIO_AG | CARBON_AG | TPA_UNADJ | CN | STATECD | COUNTYCD | bio | carbon | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=160 | |||||||||||
| int64 | int64 | int64 | float64 | float64 | float64 | int64 | int64 | int64 | float64 | float64 | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
Compute per-county summaries¶
The df dataframe now includes the state and county FIPS codes, and the (adjusted) aboveground carbon and biomass. We'll group by the geographic boundaries and sum the aboveground carbon and biomass.
%%time
result = (
df.groupby(["STATECD", "COUNTYCD"])[["bio", "carbon"]]
.sum()
.compute()
.reset_index()
.assign(
STATE=lambda df: df["STATECD"].astype("string").str.pad(2, fillchar="0"),
COUNTY=lambda df: df["COUNTYCD"].astype("string").str.pad(3, fillchar="0"),
)
.drop(columns=["STATECD", "COUNTYCD"])
)
result.head()
CPU times: user 259 ms, sys: 36.5 ms, total: 296 ms Wall time: 35.2 s
| bio | carbon | STATE | COUNTY | |
|---|---|---|---|---|
| 0 | 9130.179946 | 4565.090000 | 01 | 001 |
| 1 | 27163.826785 | 13565.771340 | 01 | 003 |
| 2 | 15567.843806 | 7774.593220 | 01 | 005 |
| 3 | 16643.533828 | 8321.766954 | 01 | 007 |
| 4 | 8742.498366 | 4364.876867 | 01 | 009 |
Plot the results¶
Now we'll make a chloropleth for the results. We just need to join in the actual geographic boundaries of the datasets, which we can get with geopandas.
census_item = catalog.get_collection("us-census").get_item(
"2020-cb_2020_us_county_500k"
)
counties = (
dask_geopandas.read_parquet(
census_item.assets["data"].href,
storage_options=census_item.assets["data"].extra_fields[
"table:storage_options"
],
columns=["STATEFP", "COUNTYFP", "geometry"],
).rename(columns={"STATEFP": "STATE", "COUNTYFP": "COUNTY"})
).compute()
Finally, we'll slice the data down to the continental United States (the dataset covers Hawaii, Alaska, and several territories).
gdf = geopandas.GeoDataFrame(pd.merge(result, counties, on=["STATE", "COUNTY"]))
df_conus = gdf.cx[-124.784:-66.951, 24.744:49.346]
df_conus.head()
| bio | carbon | STATE | COUNTY | geometry | |
|---|---|---|---|---|---|
| 0 | 9130.179946 | 4565.090000 | 01 | 001 | POLYGON ((-86.92120 32.65754, -86.92035 32.658... |
| 1 | 27163.826785 | 13565.771340 | 01 | 003 | POLYGON ((-88.02858 30.22676, -88.02399 30.230... |
| 2 | 15567.843806 | 7774.593220 | 01 | 005 | POLYGON ((-85.74803 31.61918, -85.74544 31.618... |
| 3 | 16643.533828 | 8321.766954 | 01 | 007 | POLYGON ((-87.42194 33.00338, -87.31854 33.006... |
| 4 | 8742.498366 | 4364.876867 | 01 | 009 | POLYGON ((-86.96336 33.85822, -86.95967 33.857... |
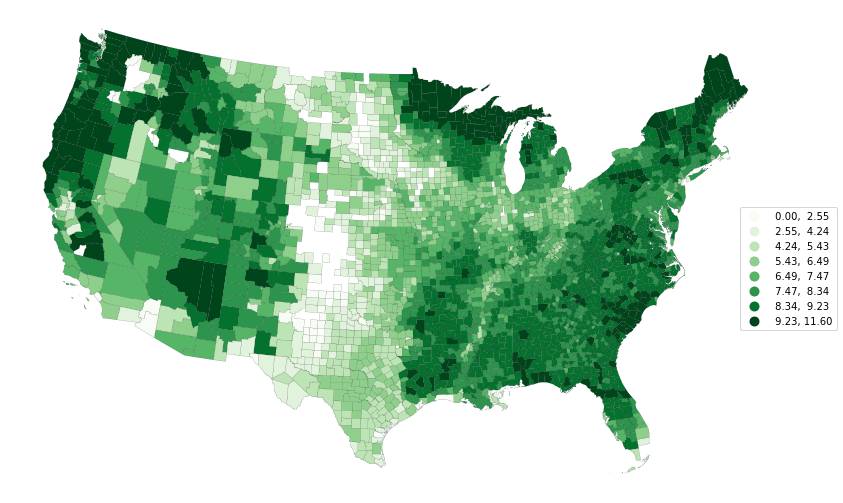
Finally, we'll plot the (log) of the estimated carbon stored above ground by the trees.
crs = ccrs.LambertConformal()
fig, ax = plt.subplots(subplot_kw={"projection": crs}, figsize=(16, 9))
df_conus.assign(carbon=np.log(df_conus.carbon + 1)).to_crs(crs.proj4_init).plot(
column="carbon",
cmap="Greens",
edgecolor="k",
scheme="natural_breaks",
k=8,
ax=ax,
linewidths=0.1,
legend=True,
)
# Shift the legend
bbox = ax.legend_.get_bbox_to_anchor().transformed(ax.transAxes.inverted())
bbox.x0 += 0.075
bbox.x1 += 0.075
bbox.y0 -= 0.4
bbox.y1 -= 0.4
ax.legend_.set_bbox_to_anchor(bbox)
ax.axis("off");
Next Steps¶
Now that you've an introduction to the Forest Inventory and Analysis dataset, learn more with
- The Reading tabular data quickstart for an introduction to tabular data on the Planeatry Computer