Accessing GBIF data with the Planetary Computer STAC API¶
This notebook provides an example of accessing Global Biodiversity Information Facility (GBIF) occurrence data from the Planetary Computer STAC API. Periodic snapshots of the data are stored in Parquet format.
import pystac_client
import planetary_computer
To access the data stored in Azure Blob Storage, we'll use the Planetary Computer's STAC API.
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
)
search = catalog.search(collections=["gbif"])
items = search.get_all_items()
items = {x.id: x for x in items}
list(items)
['gbif-2022-10-01', 'gbif-2022-09-01', 'gbif-2022-08-01', 'gbif-2022-07-01', 'gbif-2022-06-01', 'gbif-2022-05-01', 'gbif-2022-04-01', 'gbif-2022-03-01', 'gbif-2022-02-01', 'gbif-2022-01-01', 'gbif-2021-12-01', 'gbif-2021-11-01', 'gbif-2021-10-01', 'gbif-2021-09-01', 'gbif-2021-08-01', 'gbif-2021-07-01', 'gbif-2021-06-01', 'gbif-2021-04-13']
https://sasweb.microsoft.com/Member/Silo/16477We%27ll take the most recent item.
item = list(items.values())[0]
We'll use Dask to read the partitioned Parquet Dataset.
import dask.dataframe as dd
asset = item.assets["data"]
df = dd.read_parquet(
asset.href,
storage_options=asset.extra_fields["table:storage_options"],
parquet_file_extension=None,
arrow_to_pandas=dict(timestamp_as_object=True),
)
df
| gbifid | datasetkey | occurrenceid | kingdom | phylum | class | order | family | genus | species | infraspecificepithet | taxonrank | scientificname | verbatimscientificname | verbatimscientificnameauthorship | countrycode | locality | stateprovince | occurrencestatus | individualcount | publishingorgkey | decimallatitude | decimallongitude | coordinateuncertaintyinmeters | coordinateprecision | elevation | elevationaccuracy | depth | depthaccuracy | eventdate | day | month | year | taxonkey | specieskey | basisofrecord | institutioncode | collectioncode | catalognumber | recordnumber | identifiedby | dateidentified | license | rightsholder | recordedby | typestatus | establishmentmeans | lastinterpreted | mediatype | issue | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| npartitions=1960 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| int64 | object | object | object | object | object | object | object | object | object | object | object | object | object | object | object | object | object | object | int32 | object | float64 | float64 | float64 | float64 | float64 | float64 | float64 | float64 | datetime64[ns] | int32 | int32 | int32 | int32 | int32 | object | object | object | object | object | object | datetime64[ns] | object | object | object | object | object | datetime64[ns] | object | object | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
As indicated by npartitions, this Parquet dataset is made up of many individual parquet files. We can read in a specific partition with .get_partition
chunk = df.get_partition(0).compute()
chunk
| gbifid | datasetkey | occurrenceid | kingdom | phylum | class | order | family | genus | species | ... | identifiedby | dateidentified | license | rightsholder | recordedby | typestatus | establishmentmeans | lastinterpreted | mediatype | issue | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2141788029 | 4fa7b334-ce0d-4e88-aaae-2e0c138d049e | URN:catalog:CLO:EBIRD:OBS590769735 | Animalia | Chordata | Aves | Passeriformes | Polioptilidae | Polioptila | Polioptila caerulea | ... | [] | None | CC_BY_4_0 | None | [{'array_element': 'obsr233099'}] | [] | None | 2022-09-08 14:29:55.344000 | [] | [] |
| 1 | 2142511629 | 4fa7b334-ce0d-4e88-aaae-2e0c138d049e | URN:catalog:CLO:EBIRD:OBS591457037 | Animalia | Chordata | Aves | Columbiformes | Columbidae | Patagioenas | Patagioenas cayennensis | ... | [] | None | CC_BY_4_0 | None | [{'array_element': 'obsr767103'}] | [] | None | 2022-09-08 14:29:56.715000 | [] | [{'array_element': 'COORDINATE_ROUNDED'}] |
| 2 | 2126155116 | 4fa7b334-ce0d-4e88-aaae-2e0c138d049e | URN:catalog:CLO:EBIRD:OBS592106103 | Animalia | Chordata | Aves | Accipitriformes | Pandionidae | Pandion | Pandion haliaetus | ... | [] | None | CC_BY_4_0 | None | [{'array_element': 'obsr370369'}] | [] | None | 2022-09-08 14:29:58.118000 | [] | [] |
| 3 | 2143587663 | 4fa7b334-ce0d-4e88-aaae-2e0c138d049e | URN:catalog:CLO:EBIRD:OBS592831487 | Animalia | Chordata | Aves | Passeriformes | Icteridae | Quiscalus | Quiscalus quiscula | ... | [] | None | CC_BY_4_0 | None | [{'array_element': 'obsr29404'}] | [] | None | 2022-09-08 14:29:59.965000 | [] | [] |
| 4 | 2152055267 | 4fa7b334-ce0d-4e88-aaae-2e0c138d049e | URN:catalog:CLO:EBIRD:OBS593603038 | Animalia | Chordata | Aves | Passeriformes | Regulidae | Regulus | Regulus calendula | ... | [] | None | CC_BY_4_0 | None | [{'array_element': 'obsr383648'}] | [] | None | 2022-09-08 14:30:01.862000 | [] | [] |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 1947843 | 1674906857 | 9e932f70-0c61-11dd-84ce-b8a03c50a862 | urn:lsid:slu.aqua.rom.sers:ObservedProperty:14... | Animalia | Chordata | Actinopterygii | Perciformes | Percidae | Perca | Perca fluviatilis | ... | [] | None | CC0_1_0 | None | [{'array_element': 'Fiskeriverkets utredningsk... | [] | None | 2022-09-25 05:23:40.367000 | [] | [{'array_element': 'COORDINATE_ROUNDED'}] |
| 1947844 | 2012826837 | 9e932f70-0c61-11dd-84ce-b8a03c50a862 | urn:lsid:slu.aqua.rom.sers:ObservedProperty:14... | Animalia | Arthropoda | Malacostraca | Decapoda | Astacidae | Astacus | Astacus astacus | ... | [] | None | CC0_1_0 | None | [{'array_element': 'Fiskeriverkets utredningsk... | [] | None | 2022-09-25 05:23:40.367000 | [] | [{'array_element': 'COORDINATE_ROUNDED'}] |
| 1947845 | 1674906890 | 9e932f70-0c61-11dd-84ce-b8a03c50a862 | urn:lsid:slu.aqua.rom.sers:ObservedProperty:14... | Animalia | Chordata | Actinopterygii | Salmoniformes | Salmonidae | Salmo | Salmo trutta | ... | [] | None | CC0_1_0 | None | [{'array_element': 'Fiskeriverkets utredningsk... | [] | None | 2022-09-25 05:23:40.367000 | [] | [{'array_element': 'COORDINATE_ROUNDED'}] |
| 1947846 | 1675104176 | 9e932f70-0c61-11dd-84ce-b8a03c50a862 | urn:lsid:slu.aqua.rom.sers:ObservedProperty:14... | Animalia | Chordata | Actinopterygii | Salmoniformes | Salmonidae | Thymallus | Thymallus thymallus | ... | [] | None | CC0_1_0 | None | [{'array_element': 'Fiskeriverkets utredningsk... | [] | None | 2022-09-25 05:23:40.367000 | [] | [{'array_element': 'COORDINATE_ROUNDED'}] |
| 1947847 | 1674906836 | 9e932f70-0c61-11dd-84ce-b8a03c50a862 | urn:lsid:slu.aqua.rom.sers:ObservedProperty:14... | Animalia | Chordata | Actinopterygii | Perciformes | Percidae | Perca | Perca fluviatilis | ... | [] | None | CC0_1_0 | None | [{'array_element': 'Konsult'}] | [] | None | 2022-09-25 05:23:40.367000 | [] | [{'array_element': 'COORDINATE_ROUNDED'}] |
1947848 rows × 50 columns
To get a sense for the most commonly observed species, we'll group the dataset and get the count of each species.
chunk.groupby(["kingdom", "phylum", "class", "family", "genus"])[
"species"
].value_counts().sort_values(ascending=False).head(15)
kingdom phylum class family genus species
Animalia Chordata Actinopterygii Salmonidae Salmo Salmo trutta 36867
Cyprinidae Phoxinus Phoxinus phoxinus 16818
Aves Sturnidae Sturnus Sturnus vulgaris 12594
Actinopterygii Salmonidae Salmo Salmo salar 12313
Aves Passeridae Passer Passer domesticus 12047
Turdidae Turdus Turdus migratorius 12024
Corvidae Corvus Corvus brachyrhynchos 11941
Actinopterygii Cottidae Cottus Cottus gobio 11923
Aves Columbidae Zenaida Zenaida macroura 11370
Actinopterygii Lotidae Lota Lota lota 11167
Aves Cardinalidae Cardinalis Cardinalis cardinalis 10779
Anatidae Anas Anas platyrhynchos 10139
Corvidae Cyanocitta Cyanocitta cristata 10009
Emberizidae Melospiza Melospiza melodia 9357
Anatidae Branta Branta canadensis 8980
Name: species, dtype: int64
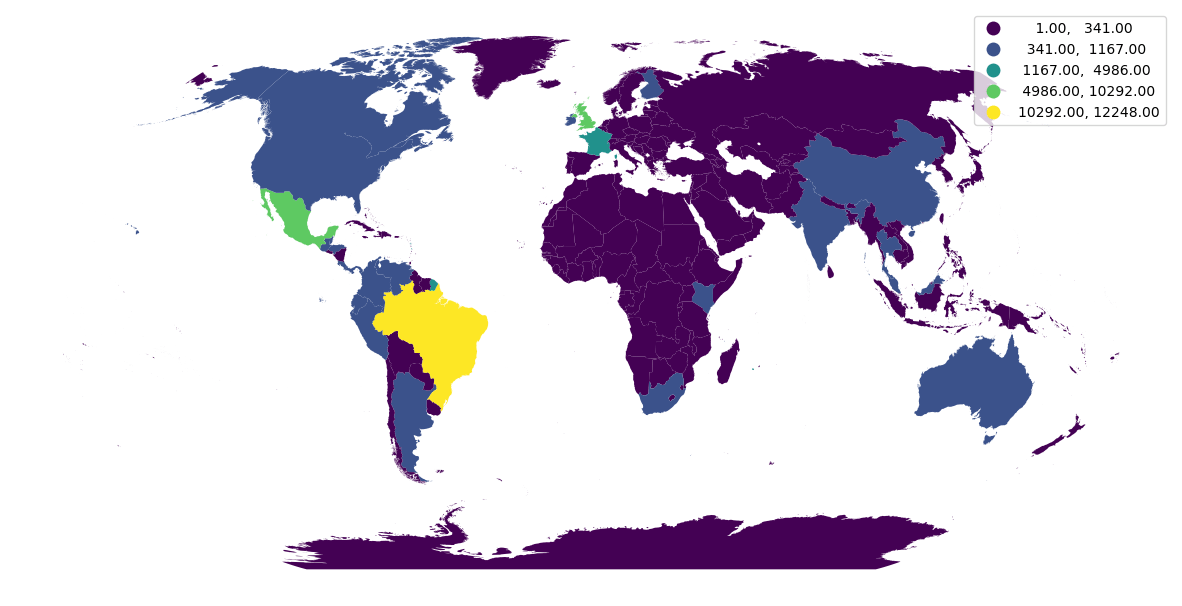
Let's create a map with the number of unique species per country. First, we'll group by country code and compute the number of unique species (per country).
species_per_country = chunk.groupby("countrycode").species.nunique()
species_per_country
countrycode
AD 4
AE 208
AF 10
AG 18
AI 11
...
YT 1
ZA 482
ZM 111
ZW 117
ZZ 15
Name: species, Length: 239, dtype: int64
Finally, we can plot the counts on a map using geopandas, by joining species_per_country to a dataset with country boundaries.
import geopandas
import cartopy
countries = geopandas.read_file(
"https://raw.githubusercontent.com/datasets/geo-countries/master/data/countries.geojson"
)
gdf = geopandas.GeoDataFrame(
countries.merge(species_per_country, left_on="ISO_A2", right_index=True)
)
crs = cartopy.crs.Robinson()
ax = gdf.to_crs(crs.proj4_init).plot(
column="species", legend=True, scheme="natural_breaks", k=5, figsize=(15, 15)
)
ax.set_axis_off()
Working with the full dataset¶
Thus far, we've just used a single partition from the full GBIF dataset. All of the examples shown in this notebook work on the entire dataset using dask.dataframe to read in the Parquet dataset.
You might want create a cluster to process the data in parallel on many machines.
from dask_gateway import GatewayCluster
cluster = GatewayCluster()
cluster.scale(16)
client = cluster.get_client()
Then use dask.dataframe.read_parquet to read in the files. To speed things up even more, we'll specify a subset of files to read in.
df = dd.read_parquet(
signed_asset.href,
columns=["countrycode", "species"],
storage_options=signed_asset.extra_fields["table:storage_options"],
)
Now you can repeat the computations above, replacing chunk with df.
Next Steps¶
Now that you've an introduction to the Forest Inventory and Analysis dataset, learn more with
- The Reading tabular data quickstart for an introduction to tabular data on the Planeatry Computer
- Scale with Dask for more on using Dask to work with large datasets