Accessing the Impact Observatory's Biodiversity Intactness data with the Planetary Computer STAC API¶
Generated by Impact Observatory, in collaboration with Vizzuality, these datasets estimate terrestrial Biodiversity Intactness as 100-meter gridded maps for the years 2017-2020. In this notebook, we'll demonstrate how to access and work with this data through the Planetary Computer.
Environment setup¶
This notebook works with or without an API key, but you will be given more permissive access to the data with an API key. If you are using the Planetary Computer Hub to run this notebook, then your API key is automatically set to the environment variable PC_SDK_SUBSCRIPTION_KEY for you when your server is started. Otherwise, you can view your keys by signing in to the developer portal. The API key may be manually set via the environment variable PC_SDK_SUBSCRIPTION_KEY or the following code:
import planetary_computer
planetary_computer.settings.set_subscription_key(<YOUR API Key>)
import pandas as pd
import planetary_computer
import pystac_client
import rich.table
import stackstac
from geogif import gif
Data access¶
The datasets hosted by the Planetary Computer are available from Azure Blob Storage. We'll use pystac-client to search the Planetary Computer's STAC API for the subset of the data that we care about, and then we'll load the data directly from Azure Blob Storage. We'll specify a modifier so that we can access the data stored in the Planetary Computer's private Blob Storage Containers. See Reading from the STAC API and Using tokens for data access for more.
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1",
modifier=planetary_computer.sign_inplace,
)
Select a region and find STAC Items¶
Let's look at an area centered on the Brazilian town of Colniza.
bbox_of_interest = [-59.958, -9.818, -58.633, -8.946]
search = catalog.search(collections=["io-biodiversity"], bbox=bbox_of_interest)
items = list(search.items())
for item in items:
print(item)
<Item id=bii_2020_-8.331364758881804_-65.13062798906854_cog> <Item id=bii_2019_-8.331364758881804_-65.13062798906854_cog> <Item id=bii_2018_-8.331364758881804_-65.13062798906854_cog> <Item id=bii_2017_-8.331364758881804_-65.13062798906854_cog>
Available Assets & Item Properties¶
Our search returned four STAC Items. We can tell from their IDs that that they contain data for the same area but for different times, specifically the years 2017 through 2020. Let's display the available assets and properties for the 2017 Item.
asset_table = rich.table.Table("Asset Key", "Asset Title")
for key, value in items[-1].assets.items():
asset_table.add_row(key, value.title)
asset_table
┏━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Asset Key ┃ Asset Title ┃ ┡━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ data │ Biodiversity Intactness │ │ tilejson │ TileJSON with default rendering │ │ rendered_preview │ Rendered preview │ └──────────────────┴─────────────────────────────────┘
property_table = rich.table.Table("Property Name", "Property Value")
for key, value in sorted(items[-1].properties.items()):
property_table.add_row(key, str(value))
property_table
┏━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Property Name ┃ Property Value ┃ ┡━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ datetime │ None │ │ end_datetime │ 2017-12-31T23:59:59Z │ │ proj:epsg │ 4326 │ │ proj:shape │ [7992, 7992] │ │ proj:transform │ [0.0008983152841195215, 0.0, -65.13062798906854, 0.0, -0.0008983152841195215, │ │ │ -8.331364758881804, 0.0, 0.0, 1.0] │ │ start_datetime │ 2017-01-01T00:00:00Z │ └────────────────┴────────────────────────────────────────────────────────────────────────────────────────────────┘
Load the Data and Plot¶
We are interested in the "data" asset. We'll use the stackstac library to read the data assets for the four Items into a single xarray.DataArray.
stack = (
stackstac.stack(items, bounds_latlon=bbox_of_interest, assets=["data"])
.assign_coords(
time=pd.to_datetime([item.properties["start_datetime"] for item in items])
.tz_convert(None)
.to_numpy()
)
.sortby("time")
)
stack.name = "Biodiversity Intactness"
stack
<xarray.DataArray 'Biodiversity Intactness' (time: 4, band: 1, y: 972, x: 1476)>
dask.array<getitem, shape=(4, 1, 972, 1476), dtype=float64, chunksize=(1, 1, 972, 1024), chunktype=numpy.ndarray>
Coordinates: (12/15)
* time (time) datetime64[ns] 2017-01-01 2018-01-01 ... 2020-01-01
id (time) <U50 'bii_2017_-8.331364758881804_-65.130627989068...
* band (band) <U4 'data'
* x (x) float64 -59.96 -59.96 -59.96 ... -58.63 -58.63 -58.63
* y (y) float64 -8.945 -8.946 -8.947 ... -9.816 -9.817 -9.818
proj:epsg int64 4326
... ...
proj:shape object {7992}
raster:bands object {'sampling': 'area', 'data_type': 'float32', 'spat...
version <U2 'v1'
title <U23 'Biodiversity Intactness'
description <U54 'Terrestrial biodiversity intactness at 100m resolut...
epsg int64 4326
Attributes:
spec: RasterSpec(epsg=4326, bounds=(-59.958053638557466, -9.818586...
crs: epsg:4326
transform: | 0.00, 0.00,-59.96|\n| 0.00,-0.00,-8.95|\n| 0.00, 0.00, 1.00|
resolution: 0.0008983152841195215At this point we haven't loaded any data into memory yet. Let's drop the single band dimension and load the data by calling .compute().
data_array = stack.squeeze().compute()
Now we can plot the data time series in a grid.
grid = data_array.plot(col="time", cmap="Greens", robust=True)
grid;
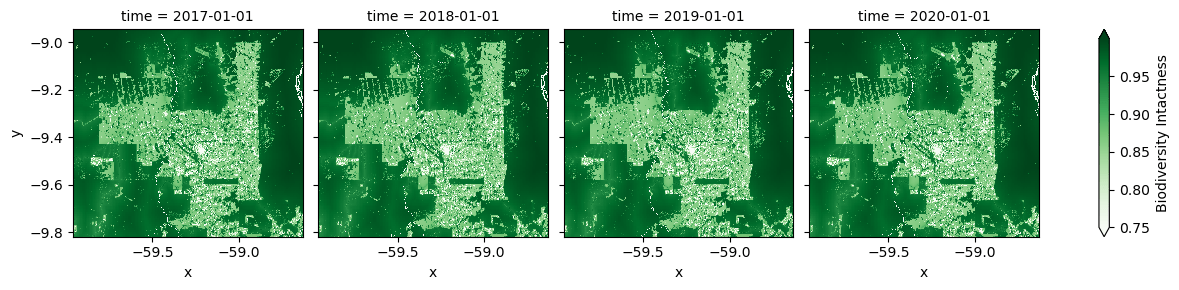
It's not easy to see the changes in a static plot, so let's make a gif that loops through the years.
gif(data_array, fps=1, cmap="Greens", robust=True)