Zonal Statistics¶
In this tutorial, you'll learn how to use xarray-spatial to work with zonal statistics. Zonal statistics help you better understand data from one source by analyzing it for different zones defined by another source. This operation uses two datasets: One dataset, the zones raster, defines one or more zones. A second dataset, the values raster, contains the data you want to analyze for each of the zones defined by the first dataset. If you're familiar with SQL or dataframe libraries like pandas, this is similar to a group by operation: you essentially group the values raster by the zones raster.
In this tutorial, we'll use the Esri / Observatory 10-Meter Land Cover dataset, which classifies pixels into one of ten classes, as the zones raster. Our values raster will be Normalized difference vegetation index (NDVI), derived from Sentinel-2 Level 2-A.
import numpy as np
import planetary_computer
import pystac_client
import stackstac
import rasterio.features
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
from xrspatial.multispectral import true_color
from xrspatial.classify import equal_interval
from xrspatial.zonal import stats as zonal_stats
from xrspatial.zonal import crosstab as zonal_crosstab
Preparation: Create a local Dask cluster¶
We'll make a local Dask cluster to process the data in parallel.
from dask.distributed import Client
client = Client()
print(f"/proxy/{client.scheduler_info()['services']['dashboard']}/status")
/proxy/8787/status
To follow the progress of your computation, you can access the Dask Dashboard at the URL from the previous cell's output.
Load and coregister data¶
The area of interest covers Lake Bridgeport, Texas, USA. We'll use pystac-client to find items covering that area, and stackstac to load the items into a DataArray.
catalog = pystac_client.Client.open(
"https://planetarycomputer.microsoft.com/api/stac/v1/",
modifier=planetary_computer.sign_inplace,
)
bounds = (-98.00080760573508, 32.99921674609716, -96.9991860639418, 34.000729644613706)
landcover_search = catalog.search(collections=["io-lulc"], bbox=bounds)
landcover_items = landcover_search.item_collection()
landcover_data = (
stackstac.stack(
landcover_items,
epsg=3857,
bounds_latlon=bounds,
dtype="int8",
fill_value=0,
chunksize=2048,
resolution=100,
)
.pipe(stackstac.mosaic, nodata=0)
.squeeze()
.rename("Landcover")
.persist()
)
landcover_data
<xarray.DataArray 'Landcover' (y: 1338, x: 1115)>
dask.array<getitem, shape=(1338, 1115), dtype=int8, chunksize=(1338, 1115), chunktype=numpy.ndarray>
Coordinates:
band <U4 'data'
* x (x) float64 -1.091e+07 -1.091e+07 ... -1.08e+07 -1.08e+07
* y (y) float64 4.029e+06 4.029e+06 ... 3.895e+06 3.895e+06
label:properties object None
label:classes object {'name': '', 'classes': ['nodata', 'water', 'tr...
label:description <U4 'lulc'
proj:shape object {74688, 101302}
label:type <U6 'raster'
start_datetime <U20 '2020-01-01T00:00:00Z'
end_datetime <U20 '2021-01-01T00:00:00Z'
raster:bands object {'nodata': 0, 'spatial_resolution': 10}
epsg int64 3857
Attributes:
spec: RasterSpec(epsg=3857, bounds=(-10909400, 3895100, -10797900,...
crs: epsg:3857
transform: | 100.00, 0.00,-10909400.00|\n| 0.00,-100.00, 4028900.00|\n|...
resolution: 100The landcover data includes some class labels, which we'll use to label the outputs later on.
landcover_labels = dict(
enumerate(landcover_data.coords["label:classes"].item()["classes"])
)
landcover_labels
{0: 'nodata',
1: 'water',
2: 'trees',
3: 'grass',
4: 'flooded veg',
5: 'crops',
6: 'scrub',
7: 'built area',
8: 'bare',
9: 'snow/ice',
10: 'clouds'}
Finally, the landcover COGs also include a colormap.
class_count = len(landcover_labels)
with rasterio.open(landcover_items[0].assets["data"].href) as src:
landcover_colormap_def = src.colormap(1) # get metadata colormap for band 1
landcover_colormap = [
np.array(landcover_colormap_def[i]) / 255 for i in range(class_count)
]
landcover_cmap = ListedColormap(landcover_colormap)
Sentinel data¶
Now we'll do the same thing, only for sentinel-2-l2a items from July 2020. We'll also try to select some not-too cloudy items and mosaic over time to remove as many clouds as possible.
sentinel_search = catalog.search(
collections=["sentinel-2-l2a"],
bbox=bounds,
datetime="2020-07-01/2020-07-30",
query={
"eo:cloud_cover": {
"lt": 10,
}
},
)
sentinel_items = sentinel_search.item_collection()
We'll need the zones and values rasters to be in the same CRS and resolution, so we'll make sure to pass those values while we're loading the data with stackstac (see Coregistration for more).
sentinel_data = (
(
stackstac.stack(
sentinel_items,
resolution=landcover_data.resolution,
epsg=landcover_data.spec.epsg,
bounds=landcover_data.spec.bounds,
assets=["B02", "B03", "B04", "B08"], # blue, green, red, nir
chunksize=2048,
)
.assign_coords(band=lambda x: x.common_name.rename("band")) # use common names
.where(lambda x: x > 0, other=np.nan) # Sentinels uses 0 as nodata
)
.median(dim="time", keep_attrs=True)
.persist()
)
sentinel_data
<xarray.DataArray 'stackstac-0446337b875acfbadb5267ecf6c67e26' (band: 4,
y: 1338, x: 1115)>
dask.array<nanmedian, shape=(4, 1338, 1115), dtype=float64, chunksize=(1, 669, 1115), chunktype=numpy.ndarray>
Coordinates: (12/19)
* band (band) <U5 'blue' 'green' ... 'nir'
* x (x) float64 -1.091e+07 ... -1.08...
* y (y) float64 4.029e+06 ... 3.895e+06
s2:datatake_type <U8 'INS-NOBS'
constellation <U10 'Sentinel 2'
sat:orbit_state <U10 'descending'
... ...
proj:shape object {10980}
title (band) <U20 'Band 2 - Blue - 10m...
common_name (band) <U5 'blue' 'green' ... 'nir'
center_wavelength (band) float64 0.49 0.56 ... 0.842
full_width_half_max (band) float64 0.098 ... 0.145
epsg int64 3857
Attributes:
spec: RasterSpec(epsg=3857, bounds=(-10909400, 3895100, -10797900,...
crs: epsg:3857
transform: | 100.00, 0.00,-10909400.00|\n| 0.00,-100.00, 4028900.00|\n|...
resolution: 100Create a true color image for the Sentinel data:
sentinel_img = true_color(
sentinel_data.sel(band="red"),
sentinel_data.sel(band="green"),
sentinel_data.sel(band="blue"),
c=30,
th=0.075,
name="True color (Sentinel)",
)
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(18, 8))
landcover_data.plot.imshow(ax=ax1, cmap=landcover_cmap)
sentinel_img.plot.imshow(ax=ax2)
plt.tight_layout()
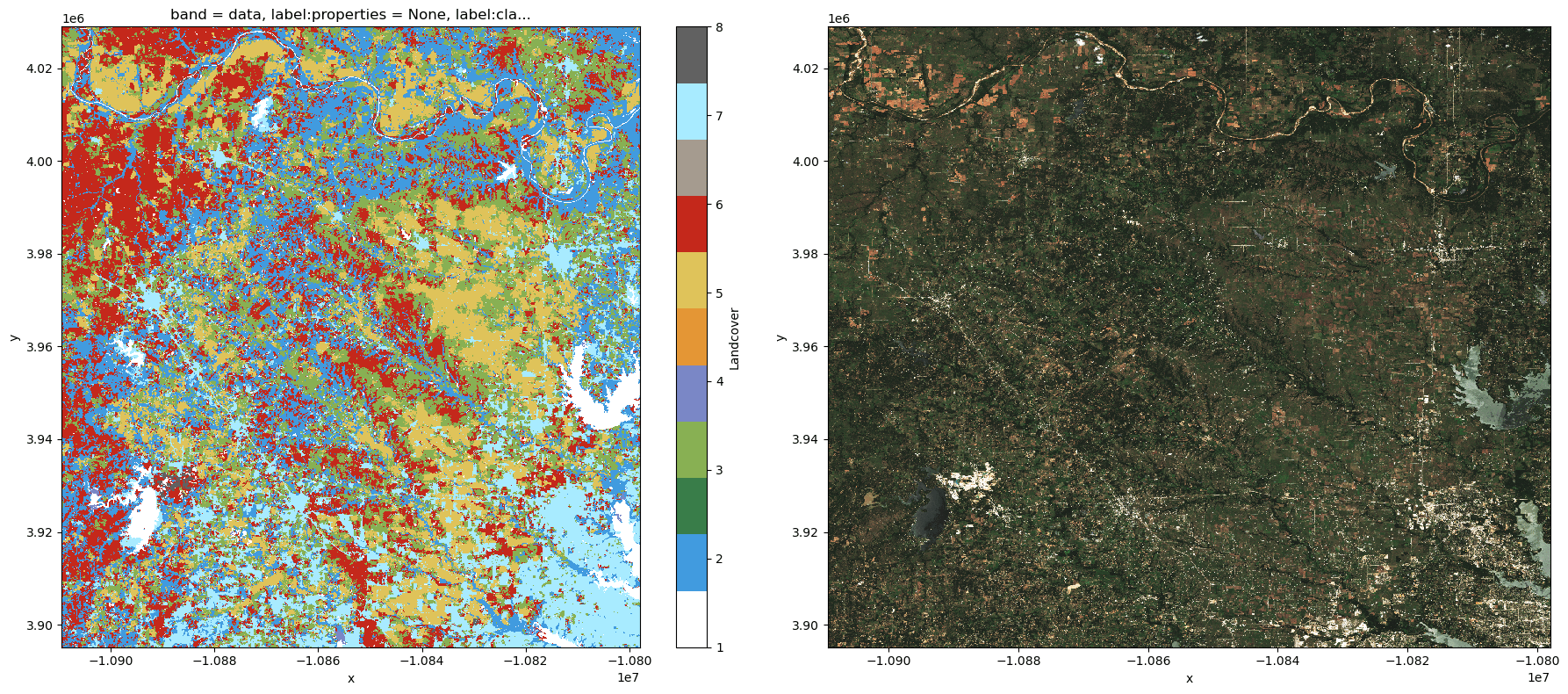
As a final bit of setup, we'll compute NDVI for the sentinel-2 scene. This will be our values raster when we compute zonal statistics later on.
red, nir = sentinel_data.sel(band=["red", "nir"])
ndvi = ((nir - red) / (nir + red)).persist()
ndvi
<xarray.DataArray 'stackstac-0446337b875acfbadb5267ecf6c67e26' (y: 1338, x: 1115)>
dask.array<truediv, shape=(1338, 1115), dtype=float64, chunksize=(669, 1115), chunktype=numpy.ndarray>
Coordinates: (12/14)
* x (x) float64 -1.091e+07 ... -1.08...
* y (y) float64 4.029e+06 ... 3.895e+06
s2:datatake_type <U8 'INS-NOBS'
constellation <U10 'Sentinel 2'
sat:orbit_state <U10 'descending'
s2:saturated_defective_pixel_percentage float64 0.0
... ...
s2:processing_baseline <U5 '02.12'
s2:degraded_msi_data_percentage float64 0.0
s2:product_type <U7 'S2MSI2A'
gsd float64 10.0
proj:shape object {10980}
epsg int64 3857Compute zonal statistics¶
Now that we have our zones raster (the landcover DataArray) and our values raster (NDVI derived from the Sentinel composite), we're ready to compute zonal stats.
We'll use xrspatial.zonal.stats to find a few summary statistics for each landcover class.
quantile_stats = (
zonal_stats(
zones=landcover_data,
values=ndvi,
stats_funcs=["mean", "max", "min", "count"],
)
.compute()
.set_index("zone")
.rename(landcover_labels)
)
quantile_stats.style.background_gradient()
/srv/conda/envs/notebook/lib/python3.10/site-packages/dask/dataframe/multi.py:1260: UserWarning: Concatenating dataframes with unknown divisions. We're assuming that the indices of each dataframes are aligned. This assumption is not generally safe. warnings.warn(
| mean | max | min | count | |
|---|---|---|---|---|
| zone | ||||
| water | 0.017884 | 0.846885 | -0.443129 | 45139.000000 |
| trees | 0.743280 | 0.900511 | -0.277344 | 339387.000000 |
| grass | 0.583749 | 0.877947 | -0.140070 | 323999.000000 |
| flooded veg | 0.547517 | 0.860692 | -0.334288 | 1573.000000 |
| crops | 0.510050 | 0.882642 | -0.252886 | 241772.000000 |
| scrub | 0.577787 | 0.879797 | -0.375246 | 336952.000000 |
| built area | 0.494027 | 0.878849 | -0.388387 | 198272.000000 |
| bare | 0.087579 | 0.704829 | -0.375067 | 4776.000000 |
Unsurprisingly, landcover classes like "trees" and "grass" have relatively high NDVI values. Classes like "water" and "bare" are relatively low.
Compute zonal cross-tabulation statistics¶
xrspatial.zonal.crosstab calculates cross-tabulated areas between the zone raster and value raster. This function requires a 2D zone raster and a categorical value raster of 2D or 3D data.
The crosstab function calculates different cross-tabulation statistics. It has an agg parameter to define which aggregation method to use. It returns a DataFrame where each row represents a zone from the zone raster and each column represents a category from the value raster.
This example uses the NASADEM elevation zones as its zone raster and the Esri Land Cover data as its categorical value raster. The resulting DataFrame will show the percentage of each land cover category for each of the five elevation zones.
nasadem_search = catalog.search(collections=["nasadem"], bbox=bounds)
nasadem_items = nasadem_search.item_collection()
nasadem_data = (
stackstac.stack(
nasadem_items,
epsg=landcover_data.spec.epsg,
resolution=landcover_data.resolution,
bounds=landcover_data.spec.bounds,
chunksize=2048,
)
.pipe(stackstac.mosaic)
.squeeze()
.persist()
).rename("Elevation (NASADEM)")
nasadem_data
<xarray.DataArray 'Elevation (NASADEM)' (y: 1338, x: 1115)>
dask.array<getitem, shape=(1338, 1115), dtype=float64, chunksize=(1338, 1115), chunktype=numpy.ndarray>
Coordinates:
band <U9 'elevation'
* x (x) float64 -1.091e+07 -1.091e+07 ... -1.08e+07 -1.08e+07
* y (y) float64 4.029e+06 4.029e+06 ... 3.895e+06 3.895e+06
proj:epsg int64 4326
proj:shape object {3601}
title <U9 'Elevation'
epsg int64 3857
Attributes:
spec: RasterSpec(epsg=3857, bounds=(-10909400, 3895100, -10797900,...
crs: epsg:3857
transform: | 100.00, 0.00,-10909400.00|\n| 0.00,-100.00, 4028900.00|\n|...
resolution: 100Now let's discretize it into five zones.
num_zones = 5
elevation_cmap = ListedColormap(plt.get_cmap("Set1").colors[:num_zones])
quantile_zones = equal_interval(
nasadem_data, k=num_zones, name="Zones (Classified Elevation - NASADEM)"
)
fig, ax = plt.subplots(figsize=(8, 8))
img = quantile_zones.plot.imshow(cmap=elevation_cmap, ax=ax)
img.colorbar.set_ticks([0.4, 1.2, 2.0, 2.8, 3.6])
img.colorbar.set_ticklabels(range(5))
ax.set_axis_off()
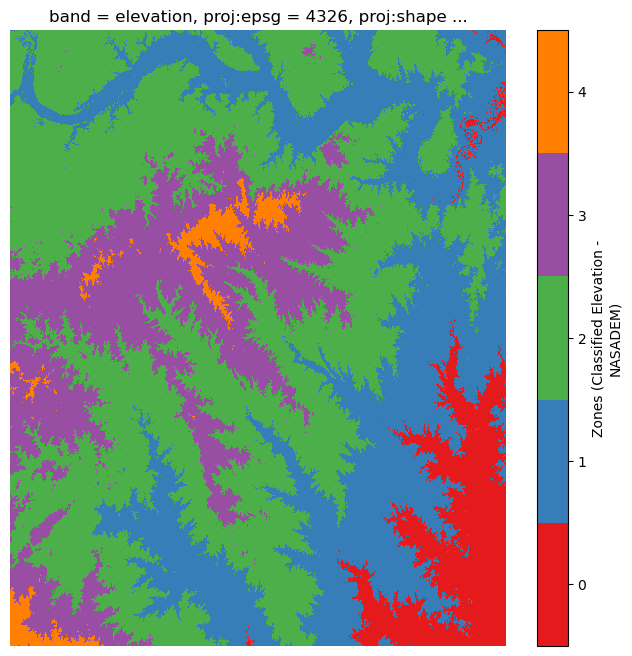
Finally, calculate the cross-tabulation statistics and display a table demonstrating how the land cover categories are distributed over each of the five elevation zones. Set values for zone_ids and cat_ids to indicate the zones and the categories of interest. In this example, we'll use all the available zones and categories.
crosstab = (
zonal_crosstab(
zones=quantile_zones,
values=landcover_data,
agg="percentage",
)
.rename(columns=landcover_labels)
.compute()
.set_index("zone")
)
crosstab.style.background_gradient()
| water | trees | grass | flooded veg | crops | scrub | built area | bare | |
|---|---|---|---|---|---|---|---|---|
| zone | ||||||||
| 0.000000 | 19.270796 | 16.616853 | 10.577962 | 0.395211 | 7.421678 | 7.088585 | 38.439863 | 0.189053 |
| 1.000000 | 2.027249 | 23.448931 | 22.147768 | 0.109607 | 20.129374 | 13.828654 | 17.774741 | 0.533675 |
| 2.000000 | 2.075033 | 20.599665 | 23.654604 | 0.090979 | 16.611000 | 27.911911 | 8.759015 | 0.297792 |
| 3.000000 | 0.596741 | 27.804037 | 21.278863 | 0.029319 | 14.013572 | 28.321761 | 7.864891 | 0.090816 |
| 4.000000 | 0.216216 | 34.702703 | 19.106518 | 0.003180 | 6.206677 | 32.282989 | 7.033386 | 0.448331 |
Next steps: analyze different datasets¶
The Planetary Computer Data Catalog includes petabytes of environmental monitoring data. All data sets are available in consistent, analysis-ready formats. You can access them through APIs as well as directly via Azure Storage.
Try using xrspatial.zonal.stats and xrspatial.zonal.crosstab with different datasets from the Data Catalog. For example, try using the land cover categories from the Esri 10m Land Cover dataset as zonal raster. Or try using the Map of Biodiversity Importance (MoBI) dataset as a values raster.
There are also other zonal functions in xarray spatial to use with your datasets.