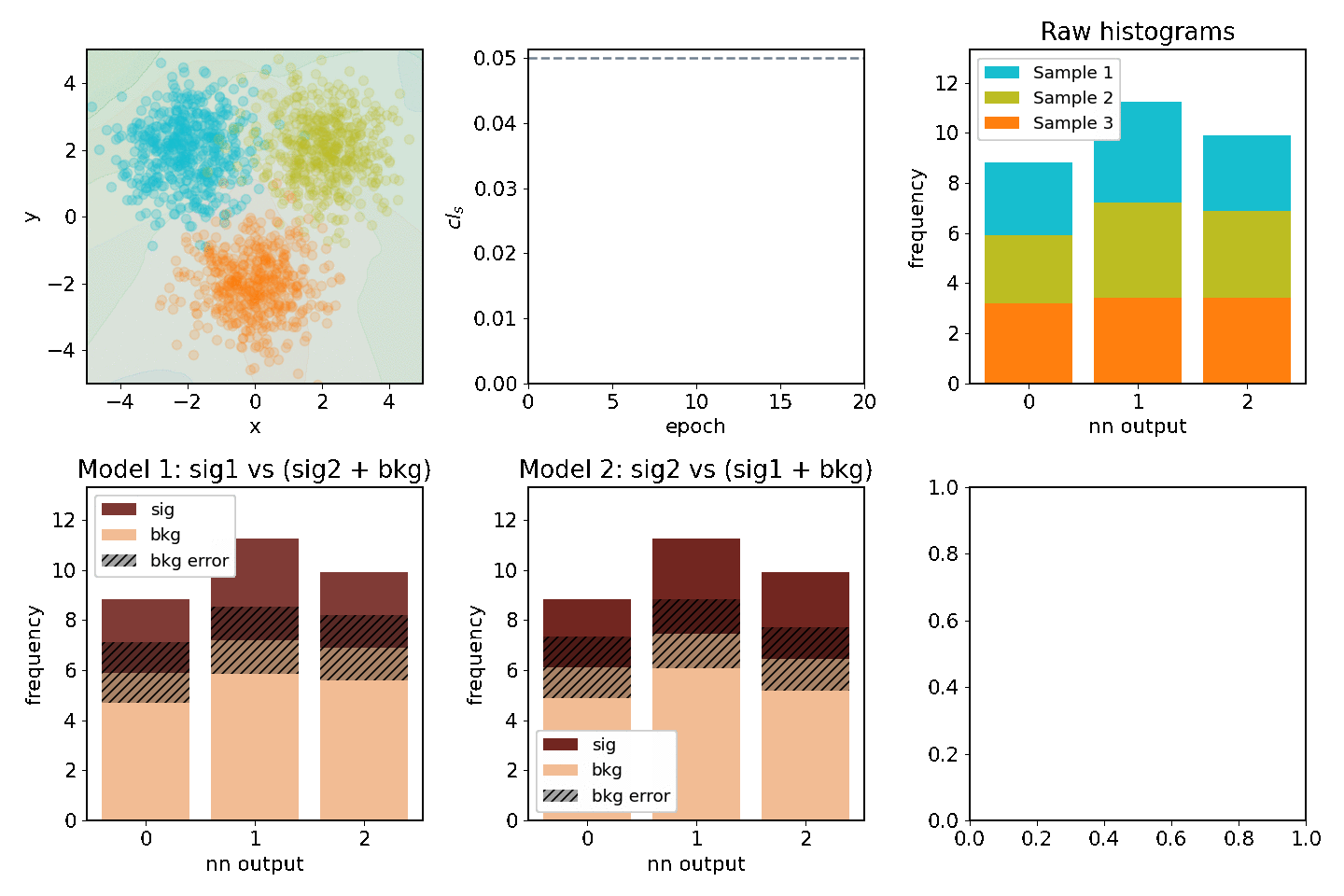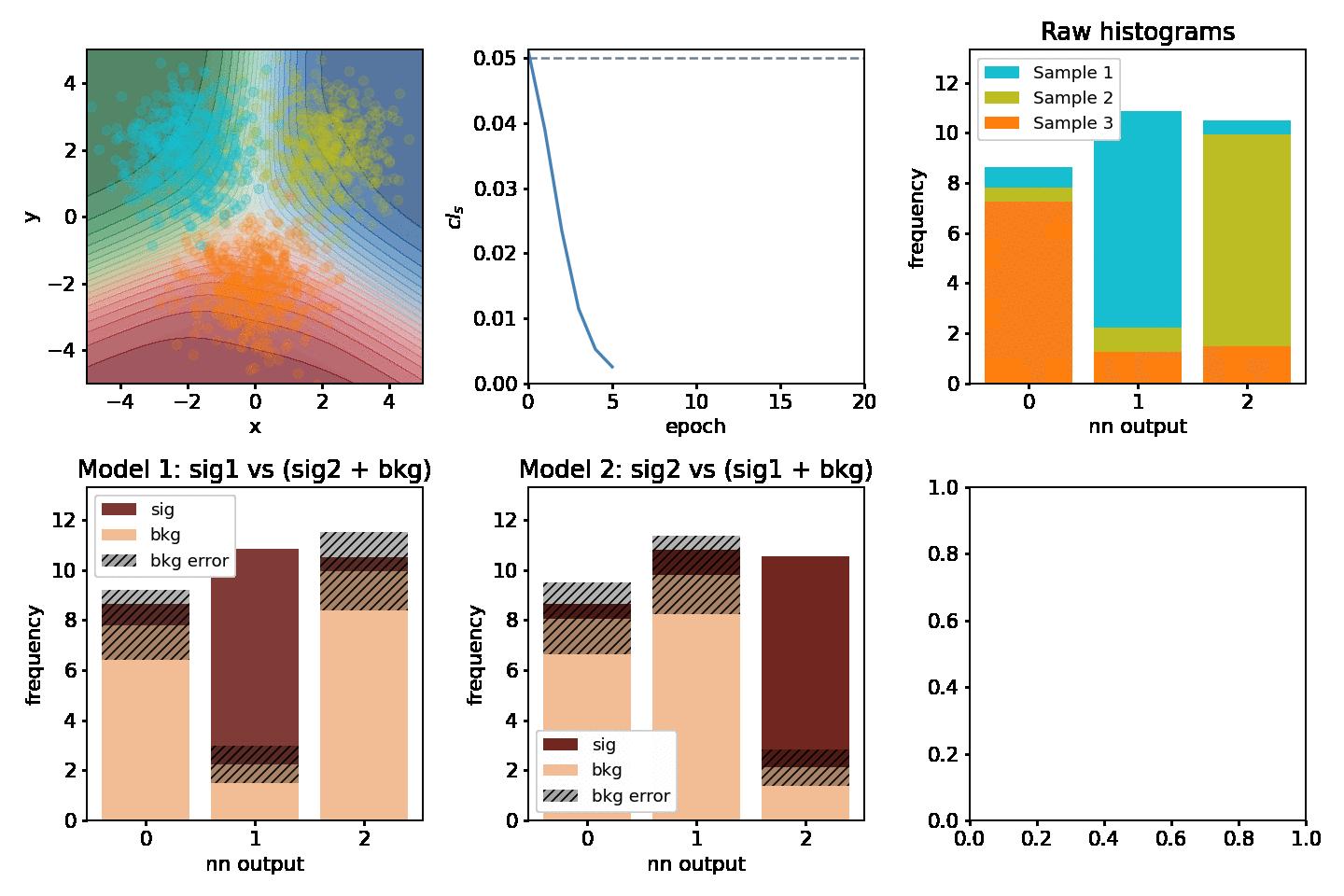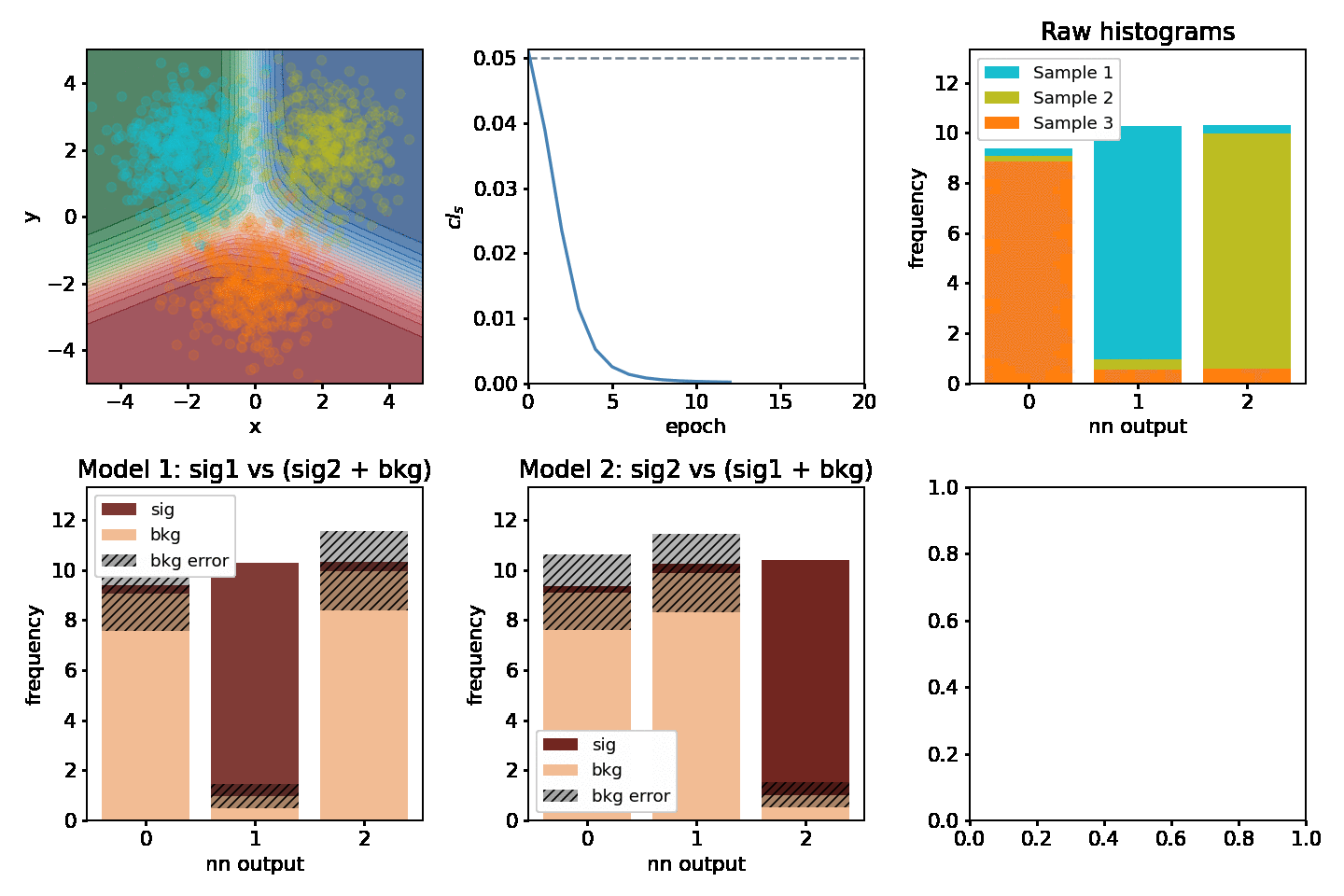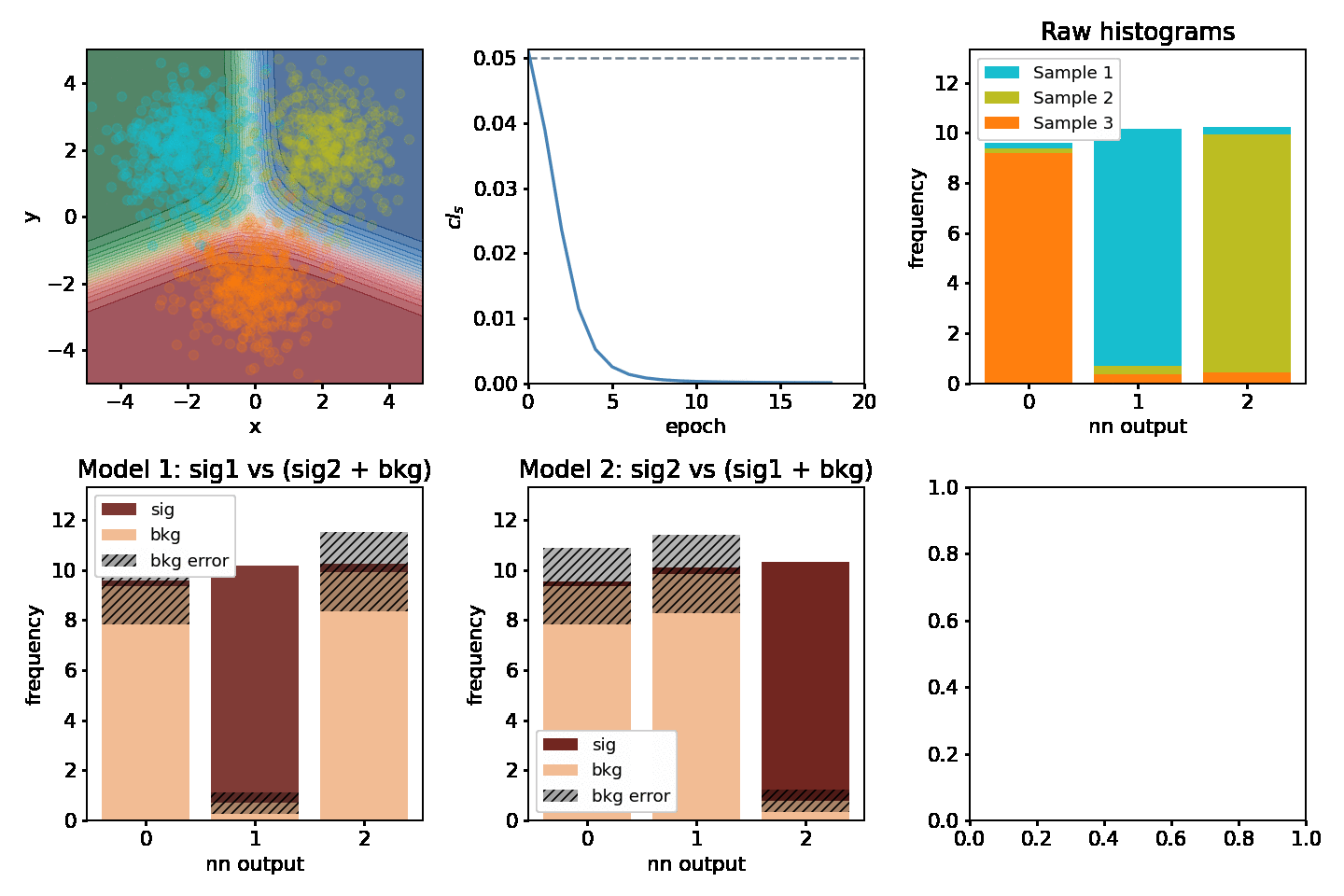
Optimizing two models at once¶
One might be interested in optimizing for two "compteting" models at the same time. Consider having 3 separate samples A, B, C and we'd be interesting in extracting the significance for two out of the three at the same time. Two models would be fitted, e.g one where A is signal and B & C are backgrounds and one where B is signal and A & C are backgrounds. This example shows how to optimize for both of them at the same time.
In [1]:
import time
import jax
import jax.experimental.optimizers as optimizers
import jax.experimental.stax as stax
import jax.random
from jax.random import PRNGKey
import numpy as np
from functools import partial
from neos import data, infer, makers
rng = PRNGKey(22)
In [2]:
def hists_from_nn_three_samples(
predict,
NMC=500,
s1_mean=[-2, 2],
s2_mean=[2, 2],
s3_mean=[0, -2],
LUMI=10,
sig_scale=1,
bkg_scale=1,
group=1,
real_z=False,
):
'''
Same as hists_from_nn_three_blobs, but parametrize grouping of signal and background for
three separatate samples
Args:
predict: Decision function for a parameterized observable. Assumed softmax here.
Returns:
hist_maker: A callable function that takes the parameters of the observable,
then constructs signal, background, and background uncertainty yields.
'''
def get_hists(network, s, bs):
NMC = len(s)
s_hist = predict(network, s).sum(axis=0) * sig_scale / NMC * LUMI
b_hists = tuple([
predict(network, bs[0]).sum(axis=0) * sig_scale / NMC * LUMI,
predict(network, bs[1]).sum(axis=0) * bkg_scale / NMC * LUMI
])
b_tot = jax.numpy.sum(jax.numpy.asarray(b_hists), axis=0)
b_unc = jax.numpy.sqrt(b_tot)
# append raw hists for signal and bkg as well
results = s_hist, b_tot, b_unc, s_hist, b_hists
return results
def hist_maker():
sig1 = np.random.multivariate_normal(s1_mean, [[1, 0], [0, 1]], size=(NMC,))
sig2 = np.random.multivariate_normal(s2_mean, [[1, 0], [0, 1]], size=(NMC,))
bkg = np.random.multivariate_normal(s3_mean, [[1, 0], [0, 1]], size=(NMC,))
def make(network):
if group == 1:
return get_hists(network, sig1, (sig2, bkg))
elif group == 2:
return get_hists(network, sig2, (sig1, bkg))
elif group == 3:
return get_hists(network, bkg, (sig1, sig2))
else:
raise UserWarning
make.bkg = bkg
make.sig2 = sig2
make.sig1 = sig1
return make
return hist_maker
In [8]:
import pyhf
pyhf.set_backend(pyhf.tensor.jax_backend())
from neos import models
def nn_hepdata_like_w_hists(histogram_maker):
'''
Analogous function to `makers.nn_hepdata_like`, but modified to pass through
the additional info added in hists_from_nn_three_samples.
'''
hm = histogram_maker()
def nn_model_maker(hpars):
network, _ = hpars
s, b, db, _, _ = hm(network) # Changed here
m = models.hepdata_like(s, b, db) # neos model
nompars = m.config.suggested_init()
bonlypars = jax.numpy.asarray([x for x in nompars])
bonlypars = jax.ops.index_update(bonlypars, m.config.poi_index, 0.0)
return m, bonlypars
nn_model_maker.hm = hm
return nn_model_maker
Initialise network using jax.experimental.stax¶
In [9]:
NOUT = 3
init_random_params, predict = stax.serial(
stax.Dense(1024),
stax.Relu,
stax.Dense(1024),
stax.Relu,
stax.Dense(NOUT),
stax.Softmax,
)
Define hitsogram and model maker functions¶
In [12]:
hmaker = hists_from_nn_three_samples(predict, group=1)
nnm = nn_hepdata_like_w_hists(hmaker)
hmaker2 = hists_from_nn_three_samples(predict, group=2)
nnm2 = nn_hepdata_like_w_hists(hmaker2)
loss1 = infer.expected_CLs(nnm, solver_kwargs=dict(pdf_transform=True))
loss2 = infer.expected_CLs(nnm2, solver_kwargs=dict(pdf_transform=True))
loss = lambda params, test_mu: (loss1(params, test_mu)[0] + loss2(params, test_mu)[0])/2
Randomly initialise nn weights and check that we can get the gradient of the loss wrt nn params¶
In [13]:
_, network = init_random_params(jax.random.PRNGKey(2), (-1, 2))
loss(network,1.)
Out[13]:
DeviceArray(0.05143672, dtype=float32)
In [14]:
nnm.hm(network)
Out[14]:
(DeviceArray([3.1857738, 3.355435 , 3.4587908], dtype=float32), DeviceArray([6.6191044, 7.0121145, 6.368781 ], dtype=float32), DeviceArray([2.572762 , 2.6480398, 2.5236444], dtype=float32), DeviceArray([3.1857738, 3.355435 , 3.4587908], dtype=float32), (DeviceArray([3.1413617, 3.741582 , 3.1170561], dtype=float32), DeviceArray([3.4777424, 3.2705328, 3.2517247], dtype=float32)))
In [15]:
a, b, c, d, e = nnm.hm(network)
Define training loop!¶
In [16]:
#jit_loss = jax.jit(loss)
opt_init, opt_update, opt_params = optimizers.adam(.5e-3)
def train_network(N, cont=False, network=None):
if not cont:
_, network = init_random_params(jax.random.PRNGKey(4), (-1, 2))
if network is not None:
network = network
losses = []
cls_vals = []
state = opt_init(network)
# parameter update function
#@jax.jit
def update_and_value(i, opt_state, mu, loss_choice):
net = opt_params(opt_state)
value, grad = jax.value_and_grad(loss_choice)(net, mu)
return opt_update(i, grad, state), value, net
for i in range(N):
start_time = time.time()
loss_choice = loss
state, value, network = update_and_value(i,state,1.0, loss_choice)
epoch_time = time.time() - start_time
losses.append(value)
metrics = {"loss": losses}
yield network, metrics, epoch_time
Plotting helper function for awesome animations :)¶
In [17]:
# Choose colormap
import matplotlib.pylab as pl
from matplotlib.colors import ListedColormap
def to_transp(cmap):
#cmap = pl.cm.Reds_r
my_cmap = cmap(np.arange(cmap.N))
#my_cmap[:,-1] = np.geomspace(0.001, 1, cmap.N)
my_cmap[:,-1] = np.linspace(0, 0.7, cmap.N)
#my_cmap[:,-1] = np.ones(cmap.N)
return ListedColormap(my_cmap)
def plot(axarr, network, metrics, hm, hm2, maxN, ith):
xlim = (-5, 5)
ylim = (-5, 5)
g = np.mgrid[xlim[0]:xlim[1]:101j, ylim[0]:ylim[1]:101j]
levels = np.linspace(0, 1, 20)
ax = axarr[0]
ax.contourf(
g[0],
g[1],
predict(network, np.moveaxis(g, 0, -1)).reshape(101, 101, NOUT)[:, :, 0],
levels=levels,
cmap = to_transp(pl.cm.Reds),
)
ax.contourf(
g[0],
g[1],
predict(network, np.moveaxis(g, 0, -1)).reshape(101, 101, NOUT)[:, :, 1],
levels=levels,
cmap = to_transp(pl.cm.Greens),
)
if NOUT > 2:
ax.contourf(
g[0],
g[1],
predict(network, np.moveaxis(g, 0, -1)).reshape(101, 101, 3)[:, :, 2],
levels=levels,
cmap = to_transp(pl.cm.Blues),
)
#print(list(map(len, [hm.sig1[:, 0], hm.sig2[:, 0], hm.bkg[:, 0]])))
ax.scatter(hm.sig1[:, 0], hm.sig1[:, 1], alpha=0.25, c="C9", label="sig1")
ax.scatter(hm.sig2[:, 0], hm.sig2[:, 1], alpha=0.17, c="C8", label="bkg2")
ax.scatter(hm.bkg[:, 0], hm.bkg[:, 1], alpha=0.17, c="C1", label="bkg2")
ax.set_xlim(*xlim)
ax.set_ylim(*ylim)
ax.set_xlabel("x")
ax.set_ylabel("y")
ax = axarr[1]
ax.axhline(0.05, c="slategray", linestyle="--")
ax.plot(metrics["loss"][:ith], c="steelblue", linewidth=2.0)
ax.set_ylim(0, metrics["loss"][0])
ax.set_xlim(0, maxN)
ax.set_xlabel("epoch")
ax.set_ylabel(r"$cl_s$")
ax = axarr[2]
s, b, db, sig, bs = hm(network)
ytop = np.max(np.sum([s, b], axis=0))*1.3
ax.bar(range(NOUT), sig, bottom=bs[0]+bs[1], color="C9", label="Sample 1")
ax.bar(range(NOUT), bs[0], bottom=bs[1], color="C8", label="Sample 2")
ax.bar(range(NOUT), bs[1], color="C1", label="Sample 3")
ax.set_ylabel("frequency")
ax.set_xlabel("nn output")
ax.set_title("Raw histograms")
ax.set_ylim(0, ytop)
if ith == 0:
ax.legend()
ax = axarr[3]
s, b, db, sig, bs = hm(network)
ax.bar(range(NOUT), s, bottom=b, color="#722620", label="sig", alpha=0.9)
ax.bar(range(NOUT), b, color="#F2BC94", label="bkg")
ax.bar(range(NOUT), db, bottom=b - db / 2.0, alpha=0.3, color="black", label="bkg error", hatch='////')
ax.set_ylabel("frequency")
ax.set_xlabel("nn output")
ax.set_title("Model 1: sig1 vs (sig2 + bkg)")
ax.set_ylim(0, ytop)
if ith == 0:
ax.legend()
ax = axarr[4]
s, b, db, sig, bs = hm2(network)
ax.bar(range(NOUT), s, bottom=b, color="#722620", label="sig")
ax.bar(range(NOUT), b, color="#F2BC94", label="bkg")
ax.bar(range(NOUT), db, bottom=b - db / 2.0, alpha=0.3, color="black", label="bkg error", hatch='////')
ax.set_ylabel("frequency")
ax.set_xlabel("nn output")
ax.set_title("Model 2: sig2 vs (sig1 + bkg)")
ax.set_ylim(0, ytop)
if ith == 0:
ax.legend()
Let's run it!!¶
In [25]:
import numpy as np
from matplotlib import pyplot as plt
from celluloid import Camera
from IPython.display import HTML
plt.rcParams.update(
{
"axes.labelsize": 13,
"axes.linewidth": 1.2,
"xtick.labelsize": 13,
"ytick.labelsize": 13,
"figure.figsize": [12.0, 8.0],
"font.size": 13,
"xtick.major.size": 3,
"ytick.major.size": 3,
"legend.fontsize": 11,
}
)
fig, axarr = plt.subplots(2, 3, dpi=120)
axarr = axarr.flatten()
# fig.set_size_inches(15, 10)
camera = Camera(fig)
maxN = 20 # make me bigger for better results!
animate = True # animations fail tests
# Training
for i, (network, metrics, epoch_time) in enumerate(train_network(maxN)):
print(f"epoch {i}:", f'CLs = {metrics["loss"][-1]}, took {epoch_time}s')
if animate:
plot(axarr, network, metrics, nnm.hm, nnm2.hm, maxN=maxN, ith=i)
plt.tight_layout()
camera.snap()
if animate:
camera.animate().save("animation.gif", writer="imagemagick", fps=10)
epoch 0: CLs = 0.051266252994537354, took 0.23153996467590332s epoch 1: CLs = 0.038961708545684814, took 0.2351398468017578s epoch 2: CLs = 0.02351313829421997, took 0.23493504524230957s epoch 3: CLs = 0.011504650115966797, took 0.23894977569580078s epoch 4: CLs = 0.005250394344329834, took 0.23163914680480957s epoch 5: CLs = 0.0025446414947509766, took 0.23407578468322754s epoch 6: CLs = 0.0013797879219055176, took 0.2348461151123047s epoch 7: CLs = 0.0008352398872375488, took 0.23739075660705566s epoch 8: CLs = 0.0005552768707275391, took 0.2395308017730713s epoch 9: CLs = 0.0003986954689025879, took 0.23309898376464844s epoch 10: CLs = 0.0003046393394470215, took 0.23505377769470215s epoch 11: CLs = 0.00024437904357910156, took 0.2402191162109375s epoch 12: CLs = 0.00020366907119750977, took 0.24132299423217773s epoch 13: CLs = 0.00017499923706054688, took 0.23680377006530762s epoch 14: CLs = 0.0001538991928100586, took 0.23770904541015625s epoch 15: CLs = 0.0001380443572998047, took 0.238480806350708s epoch 16: CLs = 0.00012576580047607422, took 0.2328932285308838s epoch 17: CLs = 0.00011610984802246094, took 0.23222708702087402s epoch 18: CLs = 0.00010830163955688477, took 0.23523879051208496s epoch 19: CLs = 0.00010198354721069336, took 0.22760796546936035s
In [ ]:
In [ ]:
In [ ]:
In [ ]: