Medical Report Generation¶
Author: Ruchit Agrawal
- Now we will demonstrate how we can use the ChexNet model to generate a medical report from an input Chest X-ray image.
Example Medical report¶

Other text reports from database¶
Comparison: two view chest from
Indication: year old male with
Findings: heart size within normal limits . mild hyperinflation of the lungs . mild pectus excavatum deformity . stable left mid lung calcified granuloma . no focal airspace disease . no pneumothorax or effusions .
Impression: changes of chronic lung disease with no acute cardiopulmonary finding .
Comparison: none .
Indication: chest ray positvie tb . language unable to get more hx
Findings: nan
Impression: normal heart size . normal pulmonary vasculature . normal mediastinal contours . lung parenchyma is clear . no airspace disease . no pulmonary edema . no of pleural effusions . no of active tuberculosis . no of active cardiopulmonary disease .
Comparison: none
Indication: chest radiograph prior to initiation of medication
Findings: the lungs appear clear . the heart and pulmonary are normal . mediastinal contours are normal . the pleural spaces are clear .
Impression: no acute cardiopulmonary disease .
from google.colab import drive
drive.mount('/content/drive')
Mounted at /content/drive
image_folder = '/content/drive/My Drive/Medical image Reporting/data/images/'
labels_file = '/content/drive/My Drive/Medical image Reporting/data/labels.csv'
chexnet_weights = '/content/drive/My Drive/Medical image Reporting/ChexNet weights/brucechou1983_CheXNet_Keras_0.3.0_weights.h5'
chexnet_weights_retrained = '/content/drive/My Drive/Medical image Reporting/ChexNet weights/chexnet_retrained-14x14.h5'
class_name = "Atelectasis,Cardiomegaly,Effusion,Infiltration,Mass,Nodule,Pneumonia,Pneumothorax,Consolidation,Edema,Emphysema,Fibrosis,Pleural_Thickening,Hernia".split(',')
import warnings
warnings.filterwarnings('ignore')
import joblib
import os
import tensorflow as tf
from tensorflow.keras.layers import Dense,GlobalAveragePooling2D, Input, Embedding, LSTM, Dot, Reshape, Concatenate, BatchNormalization, GlobalMaxPooling2D, Dropout, Add, MaxPooling2D, GRU, AveragePooling2D, Activation
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import pandas as pd
import numpy as np
import cv2
import random
import matplotlib.pyplot as plt
import matplotlib.cm as cm
import seaborn as sns
sns.set_context('notebook')
from tqdm.notebook import tqdm
from nltk.translate.bleu_score import sentence_bleu
def create_chexnet(chexnet_weights = chexnet_weights_retrained, input_size=(224,224)):
"""
Creating the CheXNet model and loading the pre-trained weights
"""
model = tf.keras.applications.DenseNet121(include_top=False, input_shape = input_size+(3,)) #importing densenet the last layer will be a relu activation layer
x = model.output #output from chexnet
x = GlobalAveragePooling2D()(x)
x = Dense(14, activation="softmax", name="chexnet_output")(x)
# x = Activation('softmax')(x)
chexnet = tf.keras.Model(inputs = model.input, outputs = x)
chexnet.load_weights(chexnet_weights)
_chexnet = tf.keras.Model(inputs = model.input, outputs = chexnet.layers[-3].output)
return chexnet, _chexnet
def predict_chexnet(img,model, plot=False):
"""
return the name of class for image
"""
class_name = "Atelectasis,Cardiomegaly,Effusion,Infiltration,Mass,Nodule,Pneumonia,Pneumothorax,Consolidation,Edema,Emphysema,Fibrosis,Pleural_Thickening,Hernia".split(',')
pred = model(img)
if plot:
fig = plt.figure(figsize=(30,4))
x = np.array(class_name)
y = np.array(pred.numpy().reshape(-1))
plt.title('Prediction probabilities')
sns.barplot(x, y, palette="Blues_d")
plt.tight_layout()
index = tf.argmax(pred, axis=1).numpy()[0]
percentage = round(pred[0,index].numpy(),2)
return class_name[index] + f' {str(percentage)}'
Encoder from ChexNet¶
- Input: Chest X-ray images
- Output: Feature vectors
class Image_encoder(tf.keras.layers.Layer):
"""
This layer will output image backbone features after passing it through CheXNet
"""
def __init__(self,
name = "image_encoder_block"
):
super().__init__()
_, self.chexnet = create_chexnet(input_size = (224,224))
self.chexnet.trainable = False
self.avgpool = AveragePooling2D()
# for i in range(10): #the last 10 layers of chexnet will be trained
# self.chexnet.layers[-i].trainable = True
def call(self, data):
op = self.chexnet(data) #op shape: (None,7,7,1024)
op = self.avgpool(op) #op shape (None,3,3,1024)
op = tf.reshape(op,shape = (-1,op.shape[1]*op.shape[2],op.shape[3])) #op shape: (None,9,1024)
return op
def encoder(image1, image2, dense_dim, dropout_rate):
"""
Takes image1,image2
gets the final encoded vector of these
"""
#image1
im_encoder = Image_encoder()
bkfeat1 = im_encoder(image1) #shape: (None,9,1024)
# print(bkfeat1.shape, 'image1 after image encoder')
bk_dense = Dense(dense_dim,name = 'bkdense',activation = 'relu') #shape: (None,9,512)
bkfeat1 = bk_dense(bkfeat1)
# print(bkfeat1.shape, 'image encoder output from bk_dense')
#image2
_bkfeat2 = im_encoder(image2) #shape: (None,9,1024)
bkfeat2 = bk_dense(_bkfeat2) #shape: (None,9,512)
#combining image1 and image2
concat = Concatenate(axis=1)([bkfeat1,bkfeat2]) #concatenating through the second axis shape: (None,18,1024)
bn = BatchNormalization(name = "encoder_batch_norm")(concat)
dropout = Dropout(dropout_rate,name = "encoder_dropout")(bn)
# print(dropout.shape, 'encoder output')
return dropout, _bkfeat2, bkfeat2
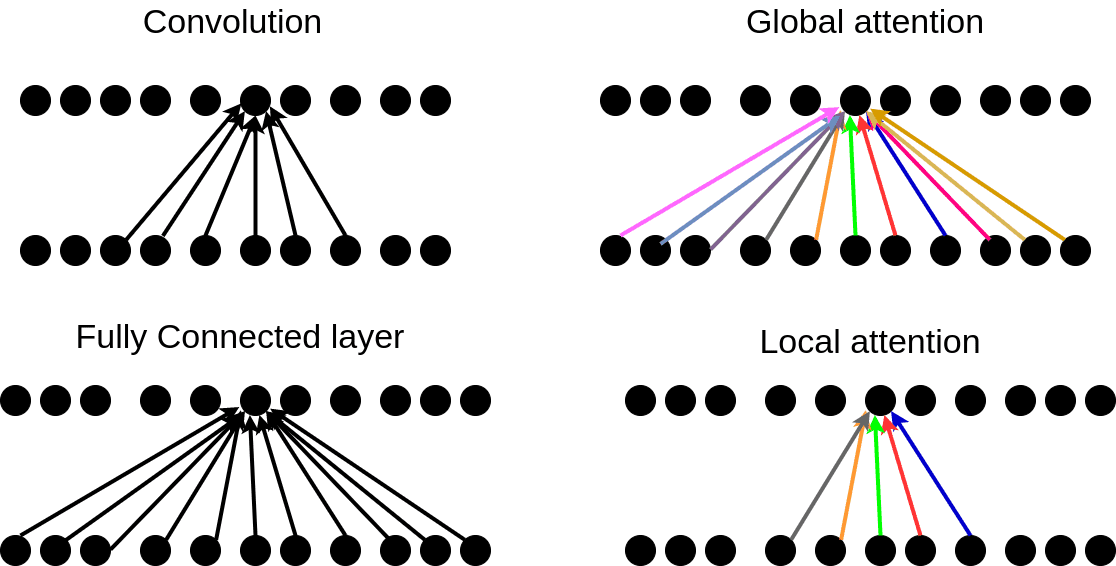
class global_attention(tf.keras.layers.Layer):
"""
calculate global attention
"""
def __init__(self, dense_dim):
super().__init__()
# Intialize variables needed for Concat score function here
self.W1 = Dense(units = dense_dim) #weight matrix of shape enc_units*dense_dim
self.W2 = Dense(units = dense_dim) #weight matrix of shape dec_units*dense_dim
self.V = Dense(units = 1) #weight matrix of shape dense_dim*1
#op (None,98,1)
def call(self,encoder_output,decoder_h): #here the encoded output will be the concatted image bk features shape: (None,98,dense_dim)
decoder_h = tf.expand_dims(decoder_h, axis=1) #shape: (None,1,dense_dim)
tanh_input = self.W1(encoder_output) + self.W2(decoder_h) #ouput_shape: batch_size*98*dense_dim
tanh_output = tf.nn.tanh(tanh_input)
attention_weights = tf.nn.softmax(self.V(tanh_output), axis=1) #shape= batch_size*98*1 getting attention alphas
op = attention_weights * encoder_output#op_shape: batch_size*98*dense_dim multiply all aplhas with corresponding context vector
context_vector = tf.reduce_sum(op, axis = 1) #summing all context vector over the time period ie input length, output_shape: batch_size*dense_dim
return context_vector,attention_weights
Decoder¶
- The decoder is based on an LSTM that takes the feature vector from the image encoder as input and generates next
hidden statesandlatent states
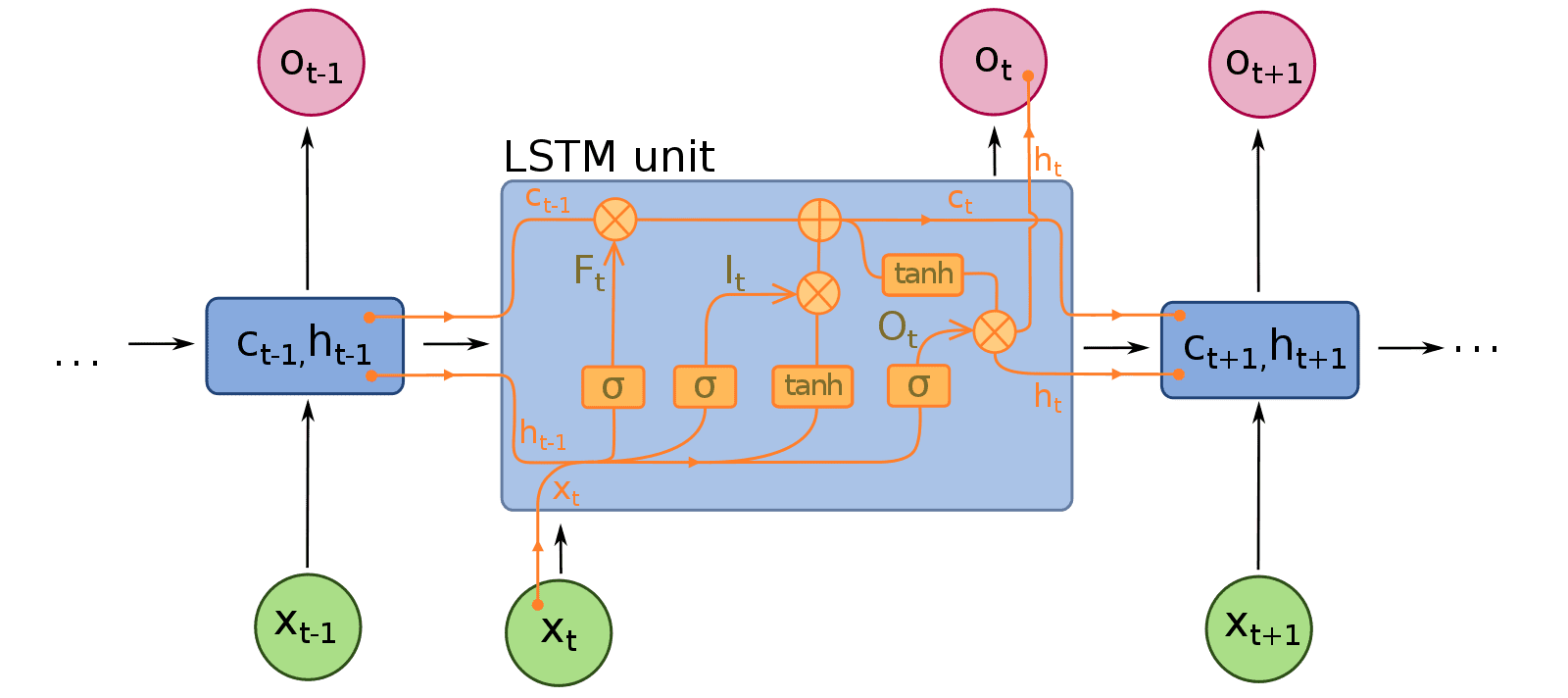
class One_Step_Decoder(tf.keras.layers.Layer):
"""
decodes a single token
"""
def __init__(self,vocab_size, embedding_dim, max_pad, dense_dim , name = "onestepdecoder"):
# Initialize decoder embedding layer, LSTM and any other objects needed
super().__init__()
self.dense_dim = dense_dim
self.embedding = Embedding(input_dim = vocab_size+1,
output_dim = embedding_dim,
input_length=max_pad,
mask_zero=True,
name = 'onestepdecoder_embedding'
)
self.LSTM = GRU(units=self.dense_dim,
# return_sequences=True,
return_state=True,
name = 'onestepdecoder_LSTM'
)
self.attention = global_attention(dense_dim = dense_dim)
self.concat = Concatenate(axis=-1)
self.dense = Dense(dense_dim, name = 'onestepdecoder_embedding_dense',activation = 'relu')
self.final = Dense(vocab_size+1, activation='softmax')
self.concat = Concatenate(axis=-1)
self.add =Add()
@tf.function
def call(self,input_to_decoder, encoder_output, decoder_h):#,decoder_c):
'''
One step decoder mechanisim step by step:
A. Pass the input_to_decoder to the embedding layer and then get the output(batch_size,1,embedding_dim)
B. Using the encoder_output and decoder hidden state, compute the context vector.
C. Concat the context vector with the step A output
D. Pass the Step-C output to LSTM/GRU and get the decoder output and states(hidden and cell state)
E. Pass the decoder output to dense layer(vocab size) and store the result into output.
F. Return the states from step D, output from Step E, attention weights from Step -B
here state_h,state_c are decoder states
'''
embedding_op = self.embedding(input_to_decoder) #output shape = batch_size*1*embedding_shape (only 1 token)
context_vector,attention_weights = self.attention(encoder_output, decoder_h) #passing hidden state h of decoder and encoder output
#context_vector shape: batch_size*dense_dim we need to add time dimension
context_vector_time_axis = tf.expand_dims(context_vector, axis=1)
#now we will combine attention output context vector with next word input to the lstm here we will be teacher forcing
concat_input = self.concat([context_vector_time_axis,embedding_op])#output dimension = batch_size*input_length(here it is 1)*(dense_dim+embedding_dim)
output,decoder_h = self.LSTM(concat_input,initial_state = decoder_h)
#output shape = batch*1*dense_dim and decoder_h,decoder_c has shape = batch*dense_dim
#we need to remove the time axis from this decoder_output
output = self.final(output)#shape = batch_size*decoder vocab size
return output,decoder_h,attention_weights
class decoder(tf.keras.Model):
"""
Decodes the encoder output and caption
"""
def __init__(self,max_pad, embedding_dim, dense_dim,batch_size ,vocab_size):
super().__init__()
self.onestepdecoder = One_Step_Decoder(vocab_size = vocab_size, embedding_dim = embedding_dim, max_pad = max_pad, dense_dim = dense_dim)
self.output_array = tf.TensorArray(tf.float32, size=max_pad)
self.max_pad = max_pad
self.batch_size = batch_size
self.dense_dim =dense_dim
@tf.function
def call(self,encoder_output,caption):#,decoder_h,decoder_c): #caption : (None,max_pad), encoder_output: (None,dense_dim)
decoder_h, decoder_c = tf.zeros_like(encoder_output[:,0]), tf.zeros_like(encoder_output[:,0]) #decoder_h, decoder_c
output_array = tf.TensorArray(tf.float32, size = self.max_pad)
for timestep in range(self.max_pad): #iterating through all timesteps ie through max_pad
output,decoder_h,attention_weights = self.onestepdecoder(caption[:,timestep:timestep+1], encoder_output, decoder_h)
output_array = output_array.write(timestep, output) #timestep*batch_size*vocab_size
self.output_array = tf.transpose(output_array.stack(),[1,0,2]) #.stack :Return the values in the TensorArray as a stacked Tensor.)
#shape output_array: (batch_size,max_pad,vocab_size)
return self.output_array, attention_weights
Putting it all together in one model¶
def create_model():
"""
creates the best model ie the attention model
and returns the model after loading the weights
and also the tokenizer
"""
#hyperparameters
input_size = (224,224)
tokenizer = joblib.load('/content/drive/MyDrive/Medical image Reporting/tokenizer.pkl')
max_pad = 29
batch_size = 100
vocab_size = len(tokenizer.word_index)
embedding_dim = 300
dense_dim = 512
lstm_units = dense_dim
dropout_rate = 0.2
tf.keras.backend.clear_session()
image1 = Input(shape = (input_size + (3,))) #shape = 224,224,3
image2 = Input(shape = (input_size + (3,))) #https://www.w3resource.com/python-exercises/tuple/python-tuple-exercise-5.php
caption = Input(shape = (max_pad,))
encoder_output, _stage1, _stage2 = encoder(image1,image2,dense_dim,dropout_rate) #shape: (None,28,512)
output,_ = decoder(max_pad, embedding_dim,dense_dim,batch_size ,vocab_size)(encoder_output,caption)
model = tf.keras.Model(inputs = [image1,image2,caption], outputs = output)
model_filename = 'Encoder_Decoder_global_attention.h5'
model_save = os.path.join('/content/drive/My Drive/Medical image Reporting', model_filename)
model.load_weights(model_save)
return model,tokenizer, [_stage1, _stage2]
def greedy_search_predict(image1, image2, model, tokenizer, input_size = (224,224)):
"""
Given paths to two x-ray images predicts the impression part of the x-ray in a greedy search algorithm
"""
# print(image1.shape, 'initial image')
image1 = tf.expand_dims(cv2.resize(image1,input_size,interpolation = cv2.INTER_NEAREST),axis=0) #introduce batch and resize
image2 = tf.expand_dims(cv2.resize(image2,input_size,interpolation = cv2.INTER_NEAREST),axis=0)
# print(image1.shape)
image1 = model.get_layer('image_encoder')(image1)
# print(image1.shape)
image2 = model.get_layer('image_encoder')(image2)
_image1 = model.get_layer('bkdense')(image1)
# print(_image1.shape)
_image2 = model.get_layer('bkdense')(image2)
concat = model.get_layer('concatenate')([_image1,_image2])
enc_op = model.get_layer('encoder_batch_norm')(concat)
enc_op = model.get_layer('encoder_dropout')(enc_op) #this is the output from encoder
# print(enc_op.shape, 'encoder output')
decoder_h,decoder_c = tf.zeros_like(enc_op[:,0]),tf.zeros_like(enc_op[:,0])
a = []
pred = []
attention_weights_list = []
max_pad = 29
for i in range(max_pad):
if i==0: #if first word
caption = np.array(tokenizer.texts_to_sequences(['<cls>'])) #shape: (1,1)
output,decoder_h,attention_weights = model.get_layer('decoder').onestepdecoder(caption,enc_op,decoder_h)#,decoder_c) decoder_c,
attention_weights_list.append(attention_weights)
#prediction
max_prob = tf.argmax(output,axis=-1) #tf.Tensor of shape = (1,1)
caption = np.array([max_prob]) #will be sent to onstepdecoder for next iteration
if max_prob==np.squeeze(tokenizer.texts_to_sequences(['<end>'])):
break;
else:
a.append(tf.squeeze(max_prob).numpy())
return tokenizer.sequences_to_texts([a])[0], [image1, _image1, attention_weights_list] #here output would be 1,1 so subscripting to open the array
def predict1(image1, image2=None, model_tokenizer = None):
"""Returns the predicted caption given the image1 and image 2 filepaths.
The model_tokenizer will contain stored model_weights and the tokenizer.
"""
if image2 == None: #if only 1 image file is given
image2 = image1
try:
image1 = cv2.imread(image1)/255
image2 = cv2.imread(image2)/255
except Exception as e:
print(e)
return print("Must be an image")
if model_tokenizer == None:
model,tokenizer, stage_list = create_model()
else:
model,tokenizer = model_tokenizer[0],model_tokenizer[1]
predicted_caption, stage_list = greedy_search_predict(image1,image2,model,tokenizer)
return predicted_caption, stage_list
def function1(image1, image2):
"""
here image1 and image2 will be a list of image
filepaths and outputs the resulting captions as a list
"""
model_tokenizer = list(create_model())
predicted_caption = []
for i1,i2 in zip(image1,image2):
caption, stages = predict1(i1,i2,model_tokenizer)
predicted_caption.append(caption)
return predicted_caption, stages
Testing the model¶
df = pd.read_csv(labels_file)
training_df_list = []
for cname in class_name:
training_df_list.append(df[df['Finding Labels']==cname].sample(14))
train_df = pd.concat(training_df_list)
train_df
| Unnamed: 0 | Image Index | Finding Labels | Follow-up # | Patient ID | Patient Age | Patient Gender | View Position | OriginalImage[Width | Height] | OriginalImagePixelSpacing[x | y] | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 150 | 434 | 00000109_000.png | Atelectasis | 0 | 109 | 54 | M | AP | 3056 | 2544 | 0.139000 | 0.139000 |
| 639 | 1987 | 00000506_023.png | Atelectasis | 23 | 506 | 27 | M | AP | 2048 | 2500 | 0.168000 | 0.168000 |
| 1275 | 4462 | 00001206_003.png | Atelectasis | 3 | 1206 | 53 | F | PA | 2048 | 2500 | 0.171000 | 0.171000 |
| 935 | 3163 | 00000830_003.png | Atelectasis | 3 | 830 | 52 | F | PA | 2021 | 2021 | 0.194311 | 0.194311 |
| 1158 | 3997 | 00001088_021.png | Atelectasis | 20 | 1088 | 45 | M | AP | 2500 | 2048 | 0.168000 | 0.168000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 5 | 9 | 00000003_006.png | Hernia | 5 | 3 | 79 | F | PA | 2992 | 2991 | 0.143000 | 0.143000 |
| 7 | 11 | 00000003_000.png | Hernia | 7 | 3 | 81 | F | PA | 2582 | 2991 | 0.143000 | 0.143000 |
| 4 | 8 | 00000003_005.png | Hernia | 4 | 3 | 78 | F | PA | 2686 | 2991 | 0.143000 | 0.143000 |
| 152 | 442 | 00000111_000.png | Hernia | 0 | 111 | 86 | F | PA | 2048 | 2500 | 0.168000 | 0.168000 |
| 2 | 5 | 00000003_002.png | Hernia | 1 | 3 | 75 | F | PA | 2048 | 2500 | 0.168000 | 0.168000 |
196 rows × 12 columns
index = random.randint(0,len(df))
name = df['Image Index'].iloc[index]
true_label = df['Finding Labels'].iloc[index]
print(name, true_label)
fig = plt.figure(figsize=(8,8))
plt.imshow(cv2.imread(image_folder+name))
plt.axis('off')
plt.tight_layout()
00000099_012.png Effusion
k = [7,300]
image1 = [image_folder+name]
result, stages = function1(image1, image1)
Output from the image encoder model (CheXNet)¶
fig = plt.figure(figsize=(32,32))
plt.imshow(stages[0][0], cmap='gray')
plt.axis('off')
plt.tight_layout()
Output after the encoded image is passed through a dense layer before being fed to the decoder¶
fig = plt.figure(figsize=(32,32))
plt.imshow(stages[1][0], cmap='gray')
plt.axis('off')
plt.tight_layout()
Attention weights over the encoded values and the hidden states of the LSTM during text generation¶
attention_img = np.zeros((stages[2][0].shape[1], stages[2][0].shape[1]))
for x in range(len(stages[2])):
attention_img[:,x] = np.array(stages[2][x][0]).reshape(-1)
fig = plt.figure(figsize=(12,12))
plt.imshow(attention_img, cmap="gray")
plt.axis('off')
plt.tight_layout()
Final Results¶
from IPython.display import display, Markdown, Latex
fig = plt.figure(figsize=(8,8))
plt.imshow(cv2.imread(image_folder+name))
plt.axis('off')
plt.tight_layout()
display(Markdown(f"## {result[0]}"))
no acute cardiopulmonary abnormality . small hiatal hernia noted .¶
indexes = random.sample(range(len(train_df)), 10)
name = train_df['Image Index'].iloc[indexes]
true_label = train_df['Finding Labels'].iloc[indexes]
fig = plt.figure(figsize=(30,30))
columns = 2
rows = 5
k = [7,300]
for i in tqdm(range(1, len(indexes)+1)):
image1 = [image_folder+name.iloc[i-1]]
result, stages = function1(image1, image1)
test_img = cv2.imread(image_folder+name.iloc[i-1])/255
fig.add_subplot(rows, columns, i)
plt.imshow(test_img, cmap='gray')
plt.text(0,-8, f" {true_label.iloc[i-1]} | gen:{result[0]}", ha='left', fontsize=14)
plt.axis('off')
plt.tight_layout()
HBox(children=(FloatProgress(value=0.0, max=10.0), HTML(value='')))