Table of Contents¶
Optimization in Julia¶
This lecture gives an overview of some optimization tools in Julia.
versioninfo()
Julia Version 1.1.0 Commit 80516ca202 (2019-01-21 21:24 UTC) Platform Info: OS: macOS (x86_64-apple-darwin14.5.0) CPU: Intel(R) Core(TM) i7-6920HQ CPU @ 2.90GHz WORD_SIZE: 64 LIBM: libopenlibm LLVM: libLLVM-6.0.1 (ORCJIT, skylake) Environment: JULIA_EDITOR = code
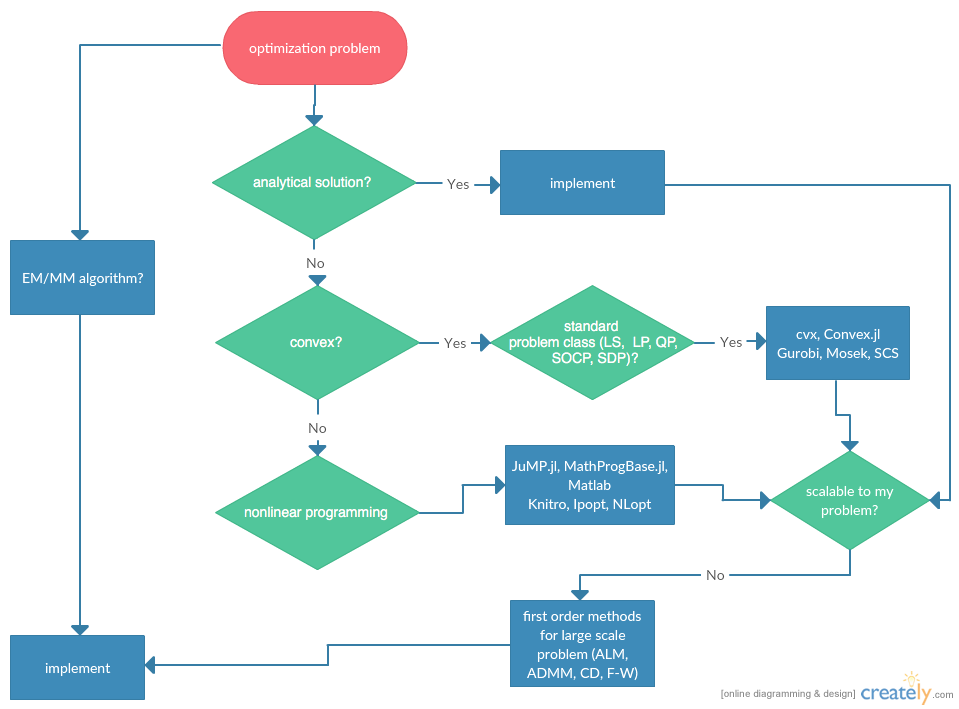
Flowchart¶
Statisticians do optimizations in daily life: maximum likelihood estimation, machine learning, ...
Category of optimization problems:
Problems with analytical solutions: least squares, principle component analysis, canonical correlation analysis, ...
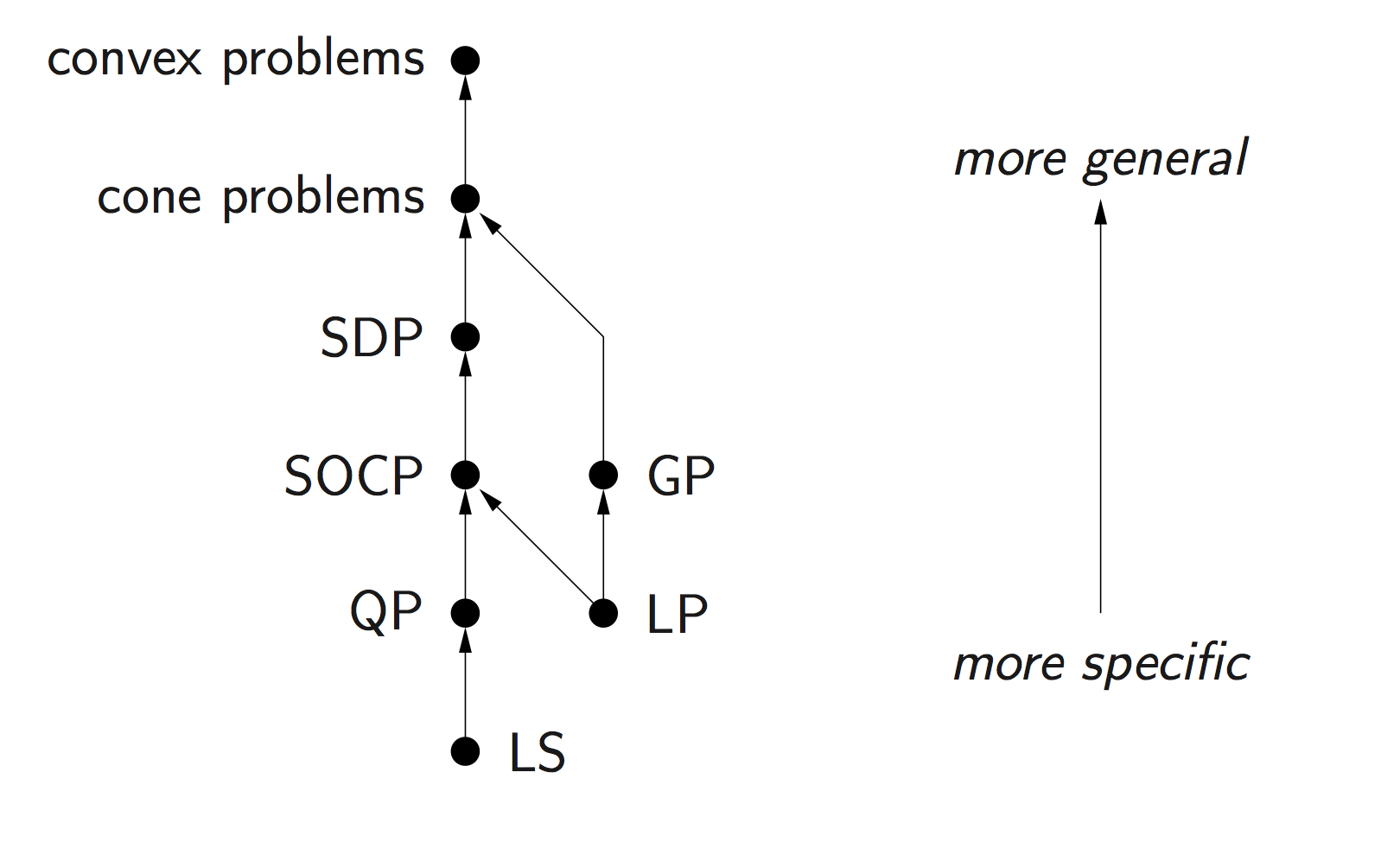
Problems subject to Disciplined Convex Programming (DCP): linear programming (LP), quadratic programming (QP), second-order cone programming (SOCP), semidefinite programming (SDP), and geometric programming (GP).
Nonlinear programming (NLP): Newton type algorithms, Fisher scoring algorithm, EM algorithm, MM algorithms.
Large scale optimization: ADMM, SGD, ...
Modeling tools and solvers¶
Getting familiar with good optimization softwares broadens the scope and scale of problems we are able to solve in statistics. Following table lists some of the best optimization softwares.
| LP | MILP | SOCP | MISOCP | SDP | GP | NLP | MINLP | R | Matlab | Julia | Python | Cost | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| modeling tools | ||||||||||||||||
| cvx | x | x | x | x | x | x | x | x | A | |||||||
| Convex.jl | x | x | x | x | x | x | O | |||||||||
| JuMP.jl | x | x | x | x | x | x | x | O | ||||||||
| MathProgBase.jl | x | x | x | x | x | x | x | O | ||||||||
| MathOptInterface.jl | x | x | x | x | x | x | x | O | ||||||||
| convex solvers | ||||||||||||||||
| Mosek | x | x | x | x | x | x | x | x | x | x | x | A | ||||
| Gurobi | x | x | x | x | x | x | x | x | A | |||||||
| CPLEX | x | x | x | x | x | x | x | x | A | |||||||
| SCS | x | x | x | x | x | x | O | |||||||||
| NLP solvers | ||||||||||||||||
| NLopt | x | x | x | x | x | O | ||||||||||
| Ipopt | x | x | x | x | x | O | ||||||||||
| KNITRO | x | x | x | x | x | x | x | x | $ |
- O: open source
- A: free academic license
- $: commercial
Difference between modeling tool and solvers
Modeling tools such as cvx (for Matlab) and Convex.jl (Julia analog of cvx) implement the disciplined convex programming (DCP) paradigm proposed by Grant and Boyd (2008) http://stanford.edu/~boyd/papers/disc_cvx_prog.html. DCP prescribes a set of simple rules from which users can construct convex optimization problems easily.
Solvers (Mosek, Gurobi, Cplex, SCS, ...) are concrete software implementation of optimization algorithms. My favorite ones are: Mosek/Gurobi/SCS for DCP and Ipopt/NLopt for nonlinear programming. Mosek and Gurobi are commercial software but free for academic use. SCS/Ipopt/NLopt are open source.
Modeling tools usually have the capability to use a variety of solvers. But modeling tools are solver agnostic so users do not have to worry about specific solver interface.
For this course, install following tools:
- Gurobi: 1. Download Gurobi at link. 2. Request free academic license at link. 3. Run
grbgetkey XXXXXXXXXcommand on terminal as suggested. It'll retrieve a license file and put it under the home folder. - Mosek: 1. Request free academic license at link. The license file will be sent to your edu email within minutes. Check Spam folder if necessary. 2. Put the license file at the default location
/home/YOURNAME/mosek/. - Convex.jl, SCS.jl, Gurobi.jl, Mosek.jl, MathProgBase.jl, NLopt.jl, Ipopt.jl.
- Gurobi: 1. Download Gurobi at link. 2. Request free academic license at link. 3. Run
DCP Using Convex.jl¶
Standard convex problem classes like LP (linear programming), QP (quadratic programming), SOCP (second-order cone programming), SDP (semidefinite programming), and GP (geometric programming), are becoming a technology.
Example: microbiome regression analysis¶
We illustrate optimization tools in Julia using microbiome analysis as an example.
16S microbiome sequencing techonology generates sequence counts of various organisms (OTUs, operational taxonomic units) in samples.
For statistical analysis, counts are normalized into proportions for each sample, resulting in a covariate matrix $\mathbf{X}$ with all rows summing to 1. For identifiability, we need to add a sum-to-zero constraint to the regression cofficients. In other words, we need to solve a constrained least squares problem
$$
\text{minimize} \frac{1}{2} \|\mathbf{y} - \mathbf{X} \beta\|_2^2
$$
subject to the constraint $\sum_{j=1}^p \beta_j = 0$. For simplicity we ignore intercept and non-OTU covariates in this presentation.
Let's first generate an artifical data set.
using Random, LinearAlgebra, SparseArrays
Random.seed!(123) # seed
n, p = 100, 50
X = rand(n, p)
lmul!(Diagonal(1 ./ vec(sum(X, dims=2))), X)
β = sprandn(p, 0.1) # sparse vector with about 10% non-zero entries
y = X * β + randn(n);
Sum-to-zero regression¶
The sum-to-zero contrained least squares is a standard quadratic programming (QP) problem so should be solved easily by any QP solver.
Modeling using Convex.jl¶
We use the Convex.jl package to model this QP problem. For a complete list of operations supported by Convex.jl, see https://convexjl.readthedocs.io/en/stable/operations.html.
using Convex
β̂cls = Variable(size(X, 2))
problem = minimize(0.5sumsquares(y - X * β̂cls)) # objective
problem.constraints += sum(β̂cls) == 0; # constraint
Mosek¶
We first use the Mosek solver to solve this QP.
using Mosek
solver = MosekSolver(LOG=1)
@time solve!(problem, solver)
Problem Name : Objective sense : min Type : CONIC (conic optimization problem) Constraints : 107 Cones : 2 Scalar variables : 157 Matrix variables : 0 Integer variables : 0 Optimizer started. Presolve started. Linear dependency checker started. Linear dependency checker terminated. Eliminator started. Freed constraints in eliminator : 0 Eliminator terminated. Eliminator - tries : 1 time : 0.00 Lin. dep. - tries : 1 time : 0.00 Lin. dep. - number : 0 Presolve terminated. Time: 0.00 Problem Name : Objective sense : min Type : CONIC (conic optimization problem) Constraints : 107 Cones : 2 Scalar variables : 157 Matrix variables : 0 Integer variables : 0 Optimizer - threads : 8 Optimizer - solved problem : the dual Optimizer - Constraints : 52 Optimizer - Cones : 3 Optimizer - Scalar variables : 106 conic : 106 Optimizer - Semi-definite variables: 0 scalarized : 0 Factor - setup time : 0.00 dense det. time : 0.00 Factor - ML order time : 0.00 GP order time : 0.00 Factor - nonzeros before factor : 1328 after factor : 1328 Factor - dense dim. : 0 flops : 3.08e+05 ITE PFEAS DFEAS GFEAS PRSTATUS POBJ DOBJ MU TIME 0 1.5e+00 7.5e-01 1.5e+00 0.00e+00 0.000000000e+00 -2.000000000e+00 1.0e+00 0.00 1 1.7e-01 8.4e-02 7.0e-02 -7.80e-01 4.353550341e+00 2.260083736e+01 1.1e-01 0.00 2 2.8e-02 1.3e-02 1.1e-02 -4.28e-01 2.091400155e+01 4.281754629e+01 1.8e-02 0.00 3 2.0e-03 9.7e-04 4.9e-03 9.83e-01 2.693910885e+01 2.746907590e+01 1.3e-03 0.00 4 1.9e-06 9.3e-07 1.2e-04 1.05e+00 2.645057050e+01 2.645136710e+01 1.2e-06 0.00 5 8.0e-10 3.9e-10 2.5e-06 1.00e+00 2.645074168e+01 2.645074202e+01 5.2e-10 0.00 Optimizer terminated. Time: 0.00 Interior-point solution summary Problem status : PRIMAL_AND_DUAL_FEASIBLE Solution status : OPTIMAL Primal. obj: 2.6450741681e+01 nrm: 5e+01 Viol. con: 8e-10 var: 0e+00 cones: 3e-08 Dual. obj: 2.6450742020e+01 nrm: 1e+01 Viol. con: 0e+00 var: 1e-08 cones: 0e+00 0.006196 seconds (3.52 k allocations: 1.643 MiB)
# Check the status, optimal value, and minimizer of the problem
problem.status, problem.optval, β̂cls.value
(:Optimal, 26.450741680579814, [20.1736; 2.11868; … ; 24.1402; -4.51783])
Gurobi¶
Switch to Gurobi solver:
using Gurobi
solver = GurobiSolver(OutputFlag=1)
@time solve!(problem, solver)
Academic license - for non-commercial use only
Optimize a model with 107 rows, 157 columns and 5160 nonzeros
Model has 2 quadratic constraints
Coefficient statistics:
Matrix range [1e-05, 2e+00]
QMatrix range [1e+00, 1e+00]
Objective range [1e+00, 1e+00]
Bounds range [0e+00, 0e+00]
RHS range [4e-03, 3e+00]
Presolve removed 2 rows and 1 columns
Presolve time: 0.00s
Presolved: 105 rows, 156 columns, 5158 nonzeros
Presolved model has 2 second-order cone constraints
Ordering time: 0.00s
Barrier statistics:
Free vars : 50
AA' NZ : 5.154e+03
Factor NZ : 5.262e+03
Factor Ops : 3.590e+05 (less than 1 second per iteration)
Threads : 1
Objective Residual
Iter Primal Dual Primal Dual Compl Time
0 1.18451802e+01 -5.01000000e-01 2.40e+01 1.00e-01 2.10e-01 0s
1 3.33648461e+00 -3.24029170e-02 4.58e+00 4.72e-05 4.06e-02 0s
2 3.12915622e+00 4.52501039e+00 3.61e+00 4.47e-05 1.99e-02 0s
3 1.51263772e+01 7.51185121e+00 1.79e+00 4.02e-05 1.02e-01 0s
4 1.38248014e+01 2.01142309e+01 1.08e+00 9.67e-05 1.68e-02 0s
5 3.30715906e+01 2.40146272e+01 3.35e-03 1.01e-05 8.59e-02 0s
6 2.71094293e+01 2.63988011e+01 3.68e-09 2.91e-06 6.71e-03 0s
7 2.64515175e+01 2.64506426e+01 1.43e-10 4.75e-08 8.22e-06 0s
8 2.64507432e+01 2.64507419e+01 5.58e-09 1.54e-09 1.39e-08 0s
Barrier solved model in 8 iterations and 0.01 seconds
Optimal objective 2.64507432e+01
0.009493 seconds (2.00 k allocations: 1.699 MiB)
# Check the status, optimal value, and minimizer of the problem
problem.status, problem.optval, β̂cls.value
(:Optimal, 26.45074321661315, [20.1748; 2.11866; … ; 24.1422; -4.51908])
Cplex¶
Switch to IBM Cplex solver:
# Use Cplex solver
using CPLEX
solver = CplexSolver(CPXPARAM_ScreenOutput=1)
@time solve!(problem, solver)
Tried aggregator 1 time. QCP Presolve eliminated 2 rows and 1 columns. Aggregator did 2 substitutions. Reduced QCP has 103 rows, 204 columns, and 5154 nonzeros. Reduced QCP has 52 quadratic constraints. Presolve time = 0.00 sec. (0.42 ticks) Parallel mode: using up to 8 threads for barrier. Number of nonzeros in lower triangle of A*A' = 5151 Using Approximate Minimum Degree ordering Total time for automatic ordering = 0.00 sec. (1.13 ticks) Summary statistics for Cholesky factor: Threads = 8 Rows in Factor = 103 Integer space required = 104 Total non-zeros in factor = 5255 Total FP ops to factor = 358959 Itn Primal Obj Dual Obj Prim Inf Upper Inf Dual Inf Inf Ratio 0 2.0710678e-01 -5.0000000e-01 8.08e+01 0.00e+00 7.30e+01 1.00e+00 1 3.8378132e+00 1.1892083e+01 8.08e+01 0.00e+00 7.30e+01 1.04e-01 2 1.4760051e+01 2.6113460e+01 7.19e+01 0.00e+00 6.50e+01 7.99e-02 3 3.2279497e+01 4.4035225e+01 5.51e+01 0.00e+00 4.98e+01 8.38e-02 4 2.7177376e+01 3.1140706e+01 8.25e+00 0.00e+00 7.46e+00 2.50e-01 5 2.6492206e+01 2.8891271e+01 1.58e+00 0.00e+00 1.43e+00 4.13e-01 6 2.6546001e+01 2.6870962e+01 9.88e-01 0.00e+00 8.93e-01 3.05e+00 7 2.6485052e+01 2.6785273e+01 1.38e-01 0.00e+00 1.24e-01 3.30e+00 8 2.6477246e+01 2.6565213e+01 1.27e-01 0.00e+00 1.15e-01 1.13e+01 9 2.6467787e+01 2.6525581e+01 3.83e-02 0.00e+00 3.46e-02 1.71e+01 10 2.6453149e+01 2.6474596e+01 2.66e-02 0.00e+00 2.41e-02 4.61e+01 11 2.6450770e+01 2.6450903e+01 1.11e-02 0.00e+00 1.00e-02 7.44e+03 12 2.6450744e+01 2.6450748e+01 6.54e-05 0.00e+00 5.91e-05 2.05e+05 13 2.6450742e+01 2.6450743e+01 2.36e-06 0.00e+00 2.14e-06 1.95e+06 0.026841 seconds (1.99 k allocations: 1.698 MiB)
# Check the status, optimal value, and minimizer of the problem
problem.status, problem.optval, β̂cls.value
(:Optimal, 26.45074204102406, [20.1739; 2.11874; … ; 24.1406; -4.51808])
SCS¶
Switch to the open source SCS solver:
# Use SCS solver
using SCS
solver = SCSSolver(verbose=1)
@time solve!(problem, solver)
0.027974 seconds (2.25 k allocations: 2.028 MiB)
----------------------------------------------------------------------------
SCS v2.0.2 - Splitting Conic Solver
(c) Brendan O'Donoghue, Stanford University, 2012-2017
----------------------------------------------------------------------------
Lin-sys: sparse-indirect, nnz in A = 5056, CG tol ~ 1/iter^(2.00)
eps = 1.00e-05, alpha = 1.50, max_iters = 5000, normalize = 1, scale = 1.00
acceleration_lookback = 20, rho_x = 1.00e-03
Variables n = 53, constraints m = 107
Cones: primal zero / dual free vars: 2
linear vars: 1
soc vars: 104, soc blks: 2
Setup time: 2.20e-04s
----------------------------------------------------------------------------
Iter | pri res | dua res | rel gap | pri obj | dua obj | kap/tau | time (s)
----------------------------------------------------------------------------
0| 4.23e+20 2.27e+20 1.00e+00 -5.02e+21 3.29e+21 8.65e+20 7.80e-04
100| 1.57e-05 1.89e-05 8.31e-07 2.64e+01 2.64e+01 1.62e-14 1.88e-02
120| 9.72e-07 4.25e-06 1.73e-09 2.65e+01 2.65e+01 5.98e-15 2.05e-02
----------------------------------------------------------------------------
Status: Solved
Timing: Solve time: 2.05e-02s
Lin-sys: avg # CG iterations: 7.60, avg solve time: 1.39e-04s
Cones: avg projection time: 2.97e-07s
Acceleration: avg step time: 2.51e-05s
----------------------------------------------------------------------------
Error metrics:
dist(s, K) = 1.6815e-11, dist(y, K*) = 1.6816e-11, s'y/|s||y| = -1.7609e-13
primal res: |Ax + s - b|_2 / (1 + |b|_2) = 9.7212e-07
dual res: |A'y + c|_2 / (1 + |c|_2) = 4.2535e-06
rel gap: |c'x + b'y| / (1 + |c'x| + |b'y|) = 1.7334e-09
----------------------------------------------------------------------------
c'x = 26.4506, -b'y = 26.4506
============================================================================
# Check the status, optimal value, and minimizer of the problem
problem.status, problem.optval, β̂cls.value
(:Optimal, 26.45062094817076, [20.1736; 2.11862; … ; 24.1401; -4.51784])
Sum-to-zero lasso¶
Suppose we want to know which organisms (OTU) are associated with the response. We can answer this question using a sum-to-zero contrained lasso $$ \text{minimize} \frac 12 \|\mathbf{y} - \mathbf{X} \beta\|_2^2 + \lambda \|\beta\|_1 $$ subject to the constraint $\sum_{j=1}^p \beta_j = 0$. Varying $\lambda$ from small to large values will generate a solution path.
# Use Mosek solver
using Mosek
solver = MosekSolver(LOG=0)
# # Use Gurobi solver
# using Gurobi
# solver = GurobiSolver(OutputFlag=0)
# Use Cplex solver
# using CPLEX
# solver = CplexSolver(CPXPARAM_ScreenOutput=0)
# # Use SCS solver
# using SCS
# solver = SCSSolver(verbose=0)
# solve at a grid of λ
λgrid = 0:0.01:0.35
β̂path = zeros(length(λgrid), size(X, 2)) # each row is β̂ at a λ
β̂classo = Variable(size(X, 2))
@time for i in 1:length(λgrid)
λ = λgrid[i]
# objective
problem = minimize(0.5sumsquares(y - X * β̂classo) + λ * sum(abs, β̂classo))
# constraint
problem.constraints += sum(β̂classo) == 0 # constraint
solve!(problem, solver)
β̂path[i, :] = β̂classo.value
end
1.364175 seconds (1.25 M allocations: 838.979 MiB, 26.14% gc time)
using Plots; gr()
using LaTeXStrings
p = plot(collect(λgrid), β̂path, legend=:none)
xlabel!(p, L"\lambda")
ylabel!(p, L"\hat \beta")
title!(p, "Sum-to-Zero Lasso")
Sum-to-zero group lasso¶
Suppose we want to do variable selection not at the OTU level, but at the Phylum level. OTUs are clustered into various Phyla. We can answer this question using a sum-to-zero contrained group lasso $$ \text{minimize} \frac 12 \|\mathbf{y} - \mathbf{X} \beta\|_2^2 + \lambda \sum_j \|\mathbf{\beta}_j\|_2 $$ subject to the constraint $\sum_{j=1}^p \beta_j = 0$, where $\mathbf{\beta}_j$ are regression coefficients corresponding to the $j$-th phylum. This is a second-order cone programming (SOCP) problem readily modeled by Convex.jl.
Let's assume each 10 contiguous OTUs belong to one Phylum.
# Use Mosek solver
using Mosek
solver = MosekSolver(LOG=0)
# # Use Gurobi solver
# using Gurobi
# solver = GurobiSolver(OutputFlag=0)
# # Use Cplex solver
# using CPLEX
# solver = CplexSolver(CPXPARAM_ScreenOutput=0)
# # Use SCS solver
# using SCS
# solver = SCSSolver(verbose=0)
# solve at a grid of λ
λgrid = 0.1:0.005:0.5
β̂pathgrp = zeros(length(λgrid), size(X, 2)) # each row is β̂ at a λ
β̂classo = Variable(size(X, 2))
@time for i in 1:length(λgrid)
λ = λgrid[i]
# loss
obj = 0.5sumsquares(y - X * β̂classo)
# group lasso penalty term
for j in 1:(size(X, 2)/10)
βj = β̂classo[(10(j-1)+1):10j]
obj = obj + λ * norm(βj)
end
problem = minimize(obj)
# constraint
problem.constraints += sum(β̂classo) == 0 # constraint
solve!(problem, solver)
β̂pathgrp[i, :] = β̂classo.value
end
0.801165 seconds (400.55 k allocations: 235.576 MiB, 10.67% gc time)
We it took Mosek <1 second to solve this seemingly hard optimization problem at 80 different $\lambda$ values.
p2 = plot(collect(λgrid), β̂pathgrp, legend=:none)
xlabel!(p2, L"\lambda")
ylabel!(p2, L"\hat \beta")
title!(p2, "Sum-to-Zero Group Lasso")
Example: matrix completion¶
Load the $128 \times 128$ Lena picture with missing pixels.
using Images
lena = load("lena128missing.png")
# convert to real matrices
Y = Float64.(lena)
128×128 Array{Float64,2}:
0.0 0.0 0.635294 0.0 … 0.0 0.0 0.627451
0.627451 0.623529 0.0 0.611765 0.0 0.0 0.388235
0.611765 0.611765 0.0 0.0 0.403922 0.219608 0.0
0.0 0.0 0.611765 0.0 0.223529 0.176471 0.192157
0.611765 0.0 0.615686 0.615686 0.0 0.0 0.0
0.0 0.0 0.0 0.619608 … 0.0 0.0 0.2
0.607843 0.0 0.623529 0.0 0.176471 0.192157 0.0
0.0 0.0 0.623529 0.0 0.0 0.0 0.215686
0.619608 0.619608 0.0 0.0 0.2 0.0 0.207843
0.0 0.0 0.635294 0.635294 0.2 0.192157 0.188235
0.635294 0.0 0.0 0.0 … 0.192157 0.180392 0.0
0.631373 0.0 0.0 0.0 0.0 0.0 0.0
0.0 0.627451 0.635294 0.666667 0.172549 0.0 0.184314
⋮ ⋱ ⋮
0.0 0.129412 0.0 0.541176 0.0 0.286275 0.0
0.14902 0.129412 0.196078 0.537255 0.345098 0.0 0.0
0.215686 0.0 0.262745 0.0 0.301961 0.0 0.207843
0.345098 0.341176 0.356863 0.513725 0.0 0.0 0.231373
0.0 0.0 0.0 0.0 … 0.0 0.243137 0.258824
0.298039 0.415686 0.458824 0.0 0.0 0.0 0.258824
0.0 0.368627 0.4 0.0 0.0 0.0 0.235294
0.0 0.0 0.34902 0.0 0.0 0.239216 0.207843
0.219608 0.0 0.0 0.0 0.0 0.0 0.2
0.0 0.219608 0.235294 0.356863 … 0.0 0.0 0.0
0.196078 0.207843 0.211765 0.0 0.0 0.270588 0.345098
0.192157 0.0 0.196078 0.309804 0.266667 0.356863 0.0
We fill out the missin pixels uisng a matrix completion technique developed by Candes and Tao $$ \text{minimize } \|\mathbf{X}\|_* $$ $$ \text{subject to } x_{ij} = y_{ij} \text{ for all observed entries } (i, j). $$ Here $\|\mathbf{M}\|_* = \sum_i \sigma_i(\mathbf{M})$ is the nuclear norm. In words we seek the matrix with minimal nuclear norm that agrees with the observed entries. This is a semidefinite programming (SDP) problem readily modeled by Convex.jl.
This example takes long because of high dimensionality.
# Use Mosek solver
using Mosek
solver = MosekSolver(LOG=1)
# Linear indices of obs. entries
obsidx = findall(Y[:] .≠ 0.0)
# Create optimization variables
X = Convex.Variable(size(Y))
# Set up optmization problem
problem = minimize(nuclearnorm(X))
problem.constraints += X[obsidx] == Y[obsidx]
# Solve the problem by calling solve
@time solve!(problem, solver)
Problem Name : Objective sense : min Type : CONIC (conic optimization problem) Constraints : 73665 Cones : 0 Scalar variables : 49153 Matrix variables : 1 Integer variables : 0 Optimizer started. Presolve started. Linear dependency checker started. Linear dependency checker terminated. Eliminator started. Freed constraints in eliminator : 0 Eliminator terminated. Eliminator - tries : 1 time : 0.00 Lin. dep. - tries : 1 time : 0.01 Lin. dep. - number : 0 Presolve terminated. Time: 0.05 Problem Name : Objective sense : min Type : CONIC (conic optimization problem) Constraints : 73665 Cones : 0 Scalar variables : 49153 Matrix variables : 1 Integer variables : 0 Optimizer - threads : 8 Optimizer - solved problem : the primal Optimizer - Constraints : 32896 Optimizer - Cones : 1 Optimizer - Scalar variables : 24769 conic : 24769 Optimizer - Semi-definite variables: 1 scalarized : 32896 Factor - setup time : 231.01 dense det. time : 0.00 Factor - ML order time : 206.00 GP order time : 0.00 Factor - nonzeros before factor : 5.41e+08 after factor : 5.41e+08 Factor - dense dim. : 2 flops : 1.19e+13 ITE PFEAS DFEAS GFEAS PRSTATUS POBJ DOBJ MU TIME 0 1.0e+00 1.0e+00 1.0e+00 0.00e+00 0.000000000e+00 0.000000000e+00 1.0e+00 231.11 1 3.7e-01 3.7e-01 2.3e-01 -9.40e-01 2.723707186e+02 2.783663417e+02 3.7e-01 358.60 2 3.1e-01 3.1e-01 5.4e-01 2.89e-01 1.669310739e+02 1.662436137e+02 3.1e-01 499.40 3 5.4e-02 5.4e-02 5.3e-01 8.69e-01 1.629170332e+02 1.627092805e+02 5.4e-02 646.21 4 4.3e-03 4.3e-03 7.1e-02 1.20e+00 1.486177530e+02 1.486148653e+02 4.3e-03 796.20 5 1.6e-03 1.6e-03 4.2e-02 1.01e+00 1.483660057e+02 1.483651653e+02 1.6e-03 944.00 6 4.5e-05 4.5e-05 2.9e-02 1.00e+00 1.479825741e+02 1.479824023e+02 4.5e-05 1105.40 7 1.4e-05 1.4e-05 5.5e-03 1.00e+00 1.479752237e+02 1.479751933e+02 1.4e-05 1254.12 8 3.5e-07 3.5e-07 2.4e-03 1.00e+00 1.479711707e+02 1.479711694e+02 3.5e-07 1416.05 9 9.0e-08 8.3e-08 3.6e-04 1.00e+00 1.479711088e+02 1.479711087e+02 8.4e-08 1559.26 10 1.6e-09 1.5e-09 9.1e-05 1.00e+00 1.479710826e+02 1.479710826e+02 1.3e-09 1728.32 Optimizer terminated. Time: 1728.63 Interior-point solution summary Problem status : PRIMAL_AND_DUAL_FEASIBLE Solution status : OPTIMAL Primal. obj: 1.4797108260e+02 nrm: 1e+02 Viol. con: 3e-09 var: 0e+00 barvar: 0e+00 Dual. obj: 1.4797108259e+02 nrm: 1e+00 Viol. con: 0e+00 var: 6e-10 barvar: 3e-09 1735.908026 seconds (18.59 M allocations: 977.941 MiB, 0.04% gc time)
colorview(Gray, X.value)
Nonlinear programming (NLP)¶
We use MLE of Gamma distribution to illustrate some rudiments of nonlinear programming (NLP) in Julia.
Let $x_1,\ldots,x_m$ be a random sample from the gamma density $$ f(x) = \Gamma(\alpha)^{-1} \beta^{\alpha} x^{\alpha-1} e^{-\beta x} $$ on $(0,\infty)$. The loglikelihood function is $$ L(\alpha, \beta) = m [- \ln \Gamma(\alpha) + \alpha \ln \beta + (\alpha - 1)\overline{\ln x} - \beta \bar x], $$ where $\overline{x} = \frac{1}{m} \sum_{i=1}^m x_i$ and $\overline{\ln x} = \frac{1}{m} \sum_{i=1}^m \ln x_i$.
using Random, Statistics, SpecialFunctions
Random.seed!(280)
function gamma_logpdf(x::Vector, α::Real, β::Real)
m = length(x)
avg = mean(x)
logavg = sum(log, x) / m
m * (- lgamma(α) + α * log(β) + (α - 1) * logavg - β * avg)
end
x = rand(5)
gamma_logpdf(x, 1.0, 1.0)
-2.7886541275400365
Many optimization algorithms involve taking derivatives of the objective function. The ForwardDiff.jl package implements automatic differentiation. For example, to compute the derivative and Hessian of the log-likelihood with data x at α=1.0 and β=1.0.
using ForwardDiff
ForwardDiff.gradient(θ -> gamma_logpdf(x, θ...), [1.0; 1.0])
2-element Array{Float64,1}:
-1.058800554530917
2.2113458724599635
ForwardDiff.hessian(θ -> gamma_logpdf(x, θ...), [1.0; 1.0])
2×2 Array{Float64,2}:
-8.22467 5.0
5.0 -5.0
Generate data:
using Distributions, Random
Random.seed!(280)
(n, p) = (1000, 2)
(α, β) = 5.0 * rand(p)
x = rand(Gamma(α, β), n)
println("True parameter values:")
println("α = ", α, ", β = ", β)
True parameter values: α = 0.5535947086407722, β = 4.637963827225865
We use JuMP.jl to define and solve our NLP problem.
using JuMP, Ipopt, NLopt
m = Model(with_optimizer(Ipopt.Optimizer, print_level=3))
# m = Model(with_optimizer(NLopt.Optimizer, algorithm=:LD_MMA))
myf(a, b) = gamma_logpdf(x, a, b)
JuMP.register(m, :myf, 2, myf, autodiff=true)
@variable(m, α >= 1e-8)
@variable(m, β >= 1e-8)
@NLobjective(m, Max, myf(α, β))
print(m)
status = JuMP.optimize!(m)
println("MLE (JuMP):")
println("α = ", α, ", β = ", β)
println("Objective value: ", JuMP.objective_value(m))
println("α = ", JuMP.value(α), ", β = ", 1 / JuMP.value(β))
println("MLE (Distribution package):")
println(fit_mle(Gamma, x))
Max myf(α, β)
Subject to
α ≥ 1.0e-8
β ≥ 1.0e-8
Total number of variables............................: 2
variables with only lower bounds: 2
variables with lower and upper bounds: 0
variables with only upper bounds: 0
Total number of equality constraints.................: 0
Total number of inequality constraints...............: 0
inequality constraints with only lower bounds: 0
inequality constraints with lower and upper bounds: 0
inequality constraints with only upper bounds: 0
Number of Iterations....: 14
(scaled) (unscaled)
Objective...............: 1.8533848152383021e+03 1.8533848152383021e+03
Dual infeasibility......: 5.2040090087569420e-09 5.2040090087569420e-09
Constraint violation....: 0.0000000000000000e+00 0.0000000000000000e+00
Complementarity.........: 9.9999999999999994e-12 9.9999999999999994e-12
Overall NLP error.......: 5.2040090087569420e-09 5.2040090087569420e-09
Number of objective function evaluations = 25
Number of objective gradient evaluations = 15
Number of equality constraint evaluations = 0
Number of inequality constraint evaluations = 0
Number of equality constraint Jacobian evaluations = 0
Number of inequality constraint Jacobian evaluations = 0
Number of Lagrangian Hessian evaluations = 0
Total CPU secs in IPOPT (w/o function evaluations) = 0.019
Total CPU secs in NLP function evaluations = 0.001
EXIT: Optimal Solution Found.
MLE (JuMP):
α = α, β = β
Objective value: -1853.384815238302
α = 0.5489317142212758, β = 4.9909639237115
MLE (Distribution package):
Gamma{Float64}(α=0.5489317142213413, θ=4.990963923701522)