versioninfo()
Julia Version 1.6.2 Commit 1b93d53fc4 (2021-07-14 15:36 UTC) Platform Info: OS: macOS (x86_64-apple-darwin18.7.0) CPU: Intel(R) Core(TM) i5-8279U CPU @ 2.40GHz WORD_SIZE: 64 LIBM: libopenlibm LLVM: libLLVM-11.0.1 (ORCJIT, skylake)
using Pkg
Pkg.activate("../..")
Pkg.status()
Activating environment at `~/Dropbox/class/M1399.000200/2021/M1399_000200-2021fall/Project.toml`
Status `~/Dropbox/class/M1399.000200/2021/M1399_000200-2021fall/Project.toml` [7d9fca2a] Arpack v0.5.3 [6e4b80f9] BenchmarkTools v1.1.4 [1e616198] COSMO v0.8.1 [f65535da] Convex v0.14.13 [a93c6f00] DataFrames v1.2.2 [31a5f54b] Debugger v0.6.8 [31c24e10] Distributions v0.24.18 [e2685f51] ECOS v0.12.3 [f6369f11] ForwardDiff v0.10.19 [28b8d3ca] GR v0.58.1 [c91e804a] Gadfly v1.3.3 [bd48cda9] GraphRecipes v0.5.7 [82e4d734] ImageIO v0.5.8 [6218d12a] ImageMagick v1.2.1 [916415d5] Images v0.24.1 [b6b21f68] Ipopt v0.7.0 [42fd0dbc] IterativeSolvers v0.9.1 [4076af6c] JuMP v0.21.9 [b51810bb] MatrixDepot v1.0.4 [1ec41992] MosekTools v0.9.4 [76087f3c] NLopt v0.6.3 [47be7bcc] ORCA v0.5.0 [a03496cd] PlotlyBase v0.4.3 [f0f68f2c] PlotlyJS v0.15.0 [91a5bcdd] Plots v1.21.2 [438e738f] PyCall v1.92.3 [d330b81b] PyPlot v2.9.0 [dca85d43] QuartzImageIO v0.7.3 [6f49c342] RCall v0.13.12 [ce6b1742] RDatasets v0.7.5 [c946c3f1] SCS v0.7.1 [276daf66] SpecialFunctions v1.6.1 [2913bbd2] StatsBase v0.33.10 [b8865327] UnicodePlots v2.0.1 [0f1e0344] WebIO v0.8.15 [8f399da3] Libdl [2f01184e] SparseArrays [10745b16] Statistics
Cholesky Decomposition¶

André-Louis Cholesky, 1875-1918.
- A basic tenet in numerical analysis:
The structure should be exploited whenever possible in solving a problem.
Common structures include: symmetry, positive (semi)definiteness, sparsity, Kronecker product, low rank, ...
LU decomposition (Gaussian Elimination) is not used in statistics very often because most of time statisticians deal with positive (semi)definite matrices.
Recall that a matrix $M$ is positive (semi)definite if
- Example: normal equation
for linear regression, the coefficient matrix $\mathbf{X}^T \mathbf{X}$ is symmetric and positive semidefinite. How to exploit this structure?
- Most of time statisticians deal with positive (semi)definite matrices.
Cholesky decomposition¶
- Theorem: Let $\mathbf{A} \in \mathbb{R}^{n \times n}$ be symmetric and positive definite. Then $\mathbf{A} = \mathbf{L} \mathbf{L}^T$, where $\mathbf{L}$ is lower triangular with positive diagonal entries and is unique.
Proof (by induction):
If $n=1$, then $0 < a = \sqrt{a}\sqrt{a}$. For $n>1$, the block equation
$$
\begin{eqnarray*}
\begin{bmatrix}
a_{11} & \mathbf{a}^T \\ \mathbf{a} & \mathbf{A}_{22}
\end{bmatrix} =
\begin{bmatrix}
\ell_{11} & \mathbf{0}_{n-1}^T \\ \mathbf{b} & \mathbf{L}_{22}
\end{bmatrix}
\begin{bmatrix}
\ell_{11} & \mathbf{b}^T \\ \mathbf{0}_{n-1} & \mathbf{L}_{22}^T
\end{bmatrix}
\end{eqnarray*}
$$
entails
$$
\begin{eqnarray*}
a_{11} &=& \ell_{11}^2 \\
\mathbf{a} &=& \ell_{11} \mathbf{b} \\
\mathbf{A}_{22} &=& \mathbf{b} \mathbf{b}^T + \mathbf{L}_{22} \mathbf{L}_{22}^T
.
\end{eqnarray*}
$$
Since $a_{11}>0$ (why?), $\ell_{11}=\sqrt{a_{11}}$ and $\mathbf{b}=a_{11}^{-1/2}\mathbf{a}$ are uniquely determined.
$\mathbf{A}_{22} - \mathbf{b} \mathbf{b}^T = \mathbf{A}_{22} - a_{11}^{-1} \mathbf{a} \mathbf{a}^T$ is positive definite of size $(n-1)\times(n-1)$ because $\mathbf{A}_{22}$ is positive definite (why? homework). By induction hypothesis, lower triangular $\mathbf{L}_{22}$ such that $\mathbf{A}_{22} - \mathbf{b} \mathbf{b}^T = \mathbf{L}_{22}^T\mathbf{L}_{22}$ exists and is unique.
- This proof is constructive and completely specifies the algorithm:
for k=1:n
for j=k+1:n
a_jk_div_a_kk = A[j, k] / A[k, k]
for i=j:n
A[i, j] -= A[i, k] * a_jk_div_a_kk # L_22
end
end
sqrt_akk = sqrt(A[k, k])
for i=k:n
A[i, k] /= sqrt_akk # l_11 and b
end
end
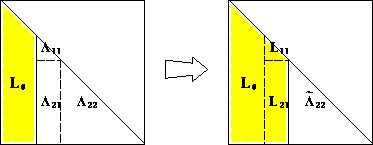
Source: http://www.netlib.org
- Computational cost:
plus $n$ square roots. Half the cost of LU decomposition.
- In general Cholesky decomposition is very stable. Failure of the decomposition simply means $\mathbf{A}$ is not positive definite. It is an efficient way to test positive definiteness.
Pivoting¶
Pivoting is only needed if you want the Cholesky factor of a positive semidefinite matrix.
When $\mathbf{A}$ does not have full rank, e.g., $\mathbf{X}^T \mathbf{X}$ with a non-full column rank $\mathbf{X}$, we encounter $a_{kk} = 0$ during the procedure.
Symmetric pivoting. At each stage $k$, we permute both row and column such that $\max_{k \le i \le n} a_{ii}$ becomes the pivot. If we encounter $\max_{k \le i \le n} a_{ii} = 0$, then $\mathbf{A}[k:n,k:n] = \mathbf{0}$ (why?) and the algorithm terminates.
With symmetric pivoting:
where $\mathbf{P}$ is a permutation matrix and $\mathbf{L} \in \mathbb{R}^{n \times r}$, $r = \text{rank}(\mathbf{A})$.
Implementation¶
Example: positive definite matrix¶
using LinearAlgebra
A = Float64.([4 12 -16; 12 37 -43; -16 -43 98]) # check out this is pd!
3×3 Matrix{Float64}:
4.0 12.0 -16.0
12.0 37.0 -43.0
-16.0 -43.0 98.0
# Cholesky without pivoting
Achol = cholesky(Symmetric(A))
Cholesky{Float64, Matrix{Float64}}
U factor:
3×3 UpperTriangular{Float64, Matrix{Float64}}:
2.0 6.0 -8.0
⋅ 1.0 5.0
⋅ ⋅ 3.0
typeof(Achol)
Cholesky{Float64, Matrix{Float64}}
fieldnames(typeof(Achol))
(:factors, :uplo, :info)
# retrieve the lower triangular Cholesky factor
Achol.L
3×3 LowerTriangular{Float64, Matrix{Float64}}:
2.0 ⋅ ⋅
6.0 1.0 ⋅
-8.0 5.0 3.0
# retrieve the upper triangular Cholesky factor
Achol.U
3×3 UpperTriangular{Float64, Matrix{Float64}}:
2.0 6.0 -8.0
⋅ 1.0 5.0
⋅ ⋅ 3.0
b = [1.0; 2.0; 3.0]
A \ b # this does LU; wasteful!; 2/3 n^3 + 2n^2
3-element Vector{Float64}:
28.58333333333338
-7.666666666666679
1.3333333333333353
Achol \ b # two triangular solves; only 2n^2 flops
3-element Vector{Float64}:
28.583333333333332
-7.666666666666666
1.3333333333333333
det(A) # this actually does LU; wasteful!
35.99999999999994
det(Achol) # cheap
36.0
inv(A) # this does LU!
3×3 Matrix{Float64}:
49.3611 -13.5556 2.11111
-13.5556 3.77778 -0.555556
2.11111 -0.555556 0.111111
inv(Achol) # 2n triangular solves
3×3 Matrix{Float64}:
49.3611 -13.5556 2.11111
-13.5556 3.77778 -0.555556
2.11111 -0.555556 0.111111
Example: positive semidefinite matrix¶
using Random
Random.seed!(123) # seed
A = randn(5, 3)
A = A * transpose(A) # A has rank 3
5×5 Matrix{Float64}:
1.97375 2.0722 1.71191 0.253774 -0.544089
2.0722 5.86947 3.01646 0.93344 -1.50292
1.71191 3.01646 2.10156 0.21341 -0.965213
0.253774 0.93344 0.21341 0.393107 -0.0415803
-0.544089 -1.50292 -0.965213 -0.0415803 0.546021
Achol = cholesky(A, Val(true)) # 2nd argument requests pivoting
RankDeficientException(1)
Stacktrace:
[1] chkfullrank
@ /Users/julia/buildbot/worker/package_macos64/build/usr/share/julia/stdlib/v1.6/LinearAlgebra/src/cholesky.jl:639 [inlined]
[2] #cholesky!#131
@ /Users/julia/buildbot/worker/package_macos64/build/usr/share/julia/stdlib/v1.6/LinearAlgebra/src/cholesky.jl:295 [inlined]
[3] cholesky!(A::Matrix{Float64}, ::Val{true}; tol::Float64, check::Bool)
@ LinearAlgebra /Users/julia/buildbot/worker/package_macos64/build/usr/share/julia/stdlib/v1.6/LinearAlgebra/src/cholesky.jl:321
[4] #cholesky#135
@ /Users/julia/buildbot/worker/package_macos64/build/usr/share/julia/stdlib/v1.6/LinearAlgebra/src/cholesky.jl:402 [inlined]
[5] cholesky(A::Matrix{Float64}, ::Val{true})
@ LinearAlgebra /Users/julia/buildbot/worker/package_macos64/build/usr/share/julia/stdlib/v1.6/LinearAlgebra/src/cholesky.jl:402
[6] top-level scope
@ In[43]:1
[7] eval
@ ./boot.jl:360 [inlined]
[8] include_string(mapexpr::typeof(REPL.softscope), mod::Module, code::String, filename::String)
@ Base ./loading.jl:1116
Achol = cholesky(A, Val(true), check=false) # turn off checking pd
CholeskyPivoted{Float64, Matrix{Float64}}
U factor with rank 4:
5×5 UpperTriangular{Float64, Matrix{Float64}}:
2.4227 0.855329 0.38529 -0.620349 1.24508
⋅ 1.11452 -0.0679895 -0.0121011 0.580476
⋅ ⋅ 0.489935 0.4013 -0.463002
⋅ ⋅ ⋅ 1.49012e-8 0.0
⋅ ⋅ ⋅ ⋅ 0.0
permutation:
5-element Vector{Int64}:
2
1
4
5
3
rank(Achol) # determine rank from Cholesky factor
4
rank(A) # determine rank from SVD, which is more numerically stable
3
Achol.L
5×5 LowerTriangular{Float64, Matrix{Float64}}:
2.4227 ⋅ ⋅ ⋅ ⋅
0.855329 1.11452 ⋅ ⋅ ⋅
0.38529 -0.0679895 0.489935 ⋅ ⋅
-0.620349 -0.0121011 0.4013 1.49012e-8 ⋅
1.24508 0.580476 -0.463002 0.0 0.0
Achol.U
5×5 UpperTriangular{Float64, Matrix{Float64}}:
2.4227 0.855329 0.38529 -0.620349 1.24508
⋅ 1.11452 -0.0679895 -0.0121011 0.580476
⋅ ⋅ 0.489935 0.4013 -0.463002
⋅ ⋅ ⋅ 1.49012e-8 0.0
⋅ ⋅ ⋅ ⋅ 0.0
Achol.p
5-element Vector{Int64}:
2
1
4
5
3
Achol.P # this returns P'
5×5 Matrix{Float64}:
0.0 1.0 0.0 0.0 0.0
1.0 0.0 0.0 0.0 0.0
0.0 0.0 0.0 0.0 1.0
0.0 0.0 1.0 0.0 0.0
0.0 0.0 0.0 1.0 0.0
# P A P' = L U
norm(transpose(Achol.P) * A * Achol.P - Achol.L * Achol.U)
2.2398766718569015e-16
Applications¶
- No inversion mentality: Whenever we see matrix inverse, we should think in terms of solving linear equations. If the matrix is positive (semi)definite, use Cholesky decomposition, which is twice cheaper than LU decomposition.
Multivariate normal density¶
Multivariate normal density $\mathcal{N}(0, \Sigma)$, where $\Sigma$ is $n\times n$ positive definite, is $$ \frac{1}{(2\pi)^{n/2}\det(\Sigma)^{1/2}}\exp\left(-\frac{1}{2}\mathbf{y}^T\Sigma^{-1}\mathbf{y}\right). $$
Hence the log likelihood is $$
- \frac{n}{2} \log (2\pi) - \frac{1}{2} \log \det \Sigma - \frac{1}{2} \mathbf{y}^T \Sigma^{-1} \mathbf{y}.
$$
Method 1:
- compute explicit inverse $\Sigma^{-1}$ ($2n^3$ flops),
- compute quadratic form ($2n^2 + 2n$ flops),
- compute determinant ($2n^3/3$ flops).
Method 2:
- Cholesky decomposition $\Sigma = \mathbf{L} \mathbf{L}^T$ ($n^3/3$ flops),
- Solve $\mathbf{L} \mathbf{x} = \mathbf{y}$ by forward substitutions ($n^2$ flops),
- compute quadratic form $\mathbf{x}^T \mathbf{x}$ ($2n$ flops)
- compute determinant from Cholesky factor ($n$ flops).
Which method is better?
# word-by-word transcription of mathematical expression
function logpdf_mvn_1(y::Vector, Σ::Matrix)
n = length(y)
- (n//2) * log(2π) - (1//2) * logdet(Σ) - (1//2) * transpose(y) * inv(Σ) * y
end
# you learnt why you should avoid inversion
function logpdf_mvn_2(y::Vector, Σ::Matrix)
n = length(y)
Σchol = cholesky(Symmetric(Σ))
- (n//2) * log(2π) - (1//2) * logdet(Σchol) - (1//2) * dot(y, Σchol \ y)
end
# this is for the efficiency-concerned
function logpdf_mvn_3(y::Vector, Σ::Matrix)
n = length(y)
Σchol = cholesky(Symmetric(Σ))
- (n//2) * log(2π) - sum(log.(diag(Σchol.L))) - (1//2) * sum(abs2, Σchol.L \ y)
end
logpdf_mvn_3 (generic function with 1 method)
using BenchmarkTools, Distributions, Random
Random.seed!(123) # seed
n = 1000
# a pd matrix
Σ = convert(Matrix{Float64}, Symmetric([i * (n - j + 1) for i in 1:n, j in 1:n]))
y = rand(MvNormal(Σ)) # one random sample from N(0, Σ)
# at least they give same answer
@show logpdf_mvn_1(y, Σ)
@show logpdf_mvn_2(y, Σ)
@show logpdf_mvn_3(y, Σ);
logpdf_mvn_1(y, Σ) = -4878.375103770505 logpdf_mvn_2(y, Σ) = -4878.375103770553 logpdf_mvn_3(y, Σ) = -4878.375103770555
@benchmark logpdf_mvn_1($y, $Σ)
BenchmarkTools.Trial: 131 samples with 1 evaluation. Range (min … max): 31.915 ms … 51.792 ms ┊ GC (min … max): 0.00% … 8.27% Time (median): 37.008 ms ┊ GC (median): 0.00% Time (mean ± σ): 38.295 ms ± 4.086 ms ┊ GC (mean ± σ): 2.88% ± 3.96% █▂▃▂ ▃▃▃▁▃▃▅▃▆▆▇████▆▇▇█▇▇▇▅▃▄▄▃▃▁▁▃▁▅▃▃▃▁▃▃▃▁▅▄▁▃▁▁▃▃▃▃▃▁▃▁▁▁▃▃ ▃ 31.9 ms Histogram: frequency by time 50.6 ms < Memory estimate: 15.78 MiB, allocs estimate: 10.
@benchmark logpdf_mvn_2($y, $Σ)
BenchmarkTools.Trial: 630 samples with 1 evaluation. Range (min … max): 5.933 ms … 18.414 ms ┊ GC (min … max): 0.00% … 0.00% Time (median): 7.527 ms ┊ GC (median): 0.00% Time (mean ± σ): 7.934 ms ± 1.511 ms ┊ GC (mean ± σ): 7.17% ± 12.28% ▂ ▃▃▁▂█▆▆▂▇▅█▃ ▂ ▄██████████████████▇▆▆▆▆▇▇▅█▅▄▄▄▄▄▅▄▄▃▃▄▆▄▃▃▃▃▂▃▃▃▃▂▁▁▁▂▂▄ ▄ 5.93 ms Histogram: frequency by time 12.5 ms < Memory estimate: 7.64 MiB, allocs estimate: 5.
@benchmark logpdf_mvn_3($y, $Σ)
BenchmarkTools.Trial: 471 samples with 1 evaluation. Range (min … max): 8.495 ms … 15.879 ms ┊ GC (min … max): 0.00% … 21.68% Time (median): 10.529 ms ┊ GC (median): 0.00% Time (mean ± σ): 10.603 ms ± 1.304 ms ┊ GC (mean ± σ): 13.33% ± 12.45% ▃▅▃▁ █▂▃ ▂▁▃▁▅▇▅▁▁▂▆▂▂▁▁ ▄ ▃ ▄▅████▇████▅▇▆▇▇███████████████▇▅█▆▇█▇▃▆▅▆▃▄▃▄▃▅▄▄▄▁▄▃▅▁▃▁▃ ▅ 8.5 ms Histogram: frequency by time 14 ms < Memory estimate: 22.91 MiB, allocs estimate: 13.
- To evaluate same multivariate normal density at many observations $y_1, y_2, \ldots$, we pre-compute the Cholesky decomposition ($n^3/3$ flops), then each evaluation costs $n^2$ flops.
Linear regression by Cholesky¶
- Cholesky decomposition is one approach to solve linear regression. Assume $\mathbf{X} \in \mathbb{R}^{n \times p}$ and $\mathbf{y} \in \mathbb{R}^n$.
- Compute $\mathbf{X}^T \mathbf{X}$: $np^2$ flops
- Compute $\mathbf{X}^T \mathbf{y}$: $2np$ flops
- Cholesky decomposition of $\mathbf{X}^T \mathbf{X}$: $\frac{1}{3} p^3$ flops
- Solve normal equation $\mathbf{X}^T \mathbf{X} \beta = \mathbf{X}^T \mathbf{y}$: $2p^2$ flops
- If need standard errors, another $(4/3)p^3$ flops
Total computational cost is $np^2 + (1/3) p^3$ (without s.e.) or $np^2 + (5/3) p^3$ (with s.e.) flops.
Further reading¶
Section 7.7 of Numerical Analysis for Statisticians of Kenneth Lange (2010).
Section II.5.3 of Computational Statistics by James Gentle (2010).
Section 4.2 of Matrix Computation by Gene Golub and Charles Van Loan (2013).
Acknowledgment¶
Many parts of this lecture note is based on Dr. Hua Zhou's 2019 Spring Statistical Computing course notes available at http://hua-zhou.github.io/teaching/biostatm280-2019spring/index.html.