Clustering with Gaussian Mixture Models¶
Preliminaries¶
- Goal
- Introduction to the Expectation-Maximization (EM) Algorithm with application to Gaussian Mixture Models (GMM)
- Materials
- Mandatory
- These lecture notes
- Optional
- Bishop pp. 430-439 for Gaussian Mixture Models
- Mandatory
Limitations of Simple IID Gaussian Models¶
Sofar, model inference was solved analytically, but we used strong assumptions:
- IID sampling, $p(D) = \prod_n p(x_n)$
- Simple Gaussian (or multinomial) PDFs, $p(x_n) = \mathcal{N}(x_n|\mu,\Sigma)$
- Some limitations of Simple Gaussian Models with IID Sampling:
- What if the PDF is multi-modal (or is just not Gaussian in any other way)?
- Covariance matrix $\Sigma$ has $D(D+1)/2$ parameters.
- This quickly becomes a very large number for increasing dimension $D$.
- Temporal signals are often not IID.
Towards More Flexible Models¶
- What if the PDF is multi-modal (or is just not Gaussian in any other way)?
- Discrete latent variable models (a.k.a. mixture models). We'll cover this case in this lesson.
- Covariance matrix $\Sigma$ has $D(D+1)/2$ parameters. This quickly becomes very large for increasing dimension $D$.
- Continuous latent variable models (a.k.a. dimensionality reduction models). Covered in lesson 11.
- Temporal signals are often not IID.
- Introduce Markov dependencies and latent state variable models. This will be covered in lesson 13.
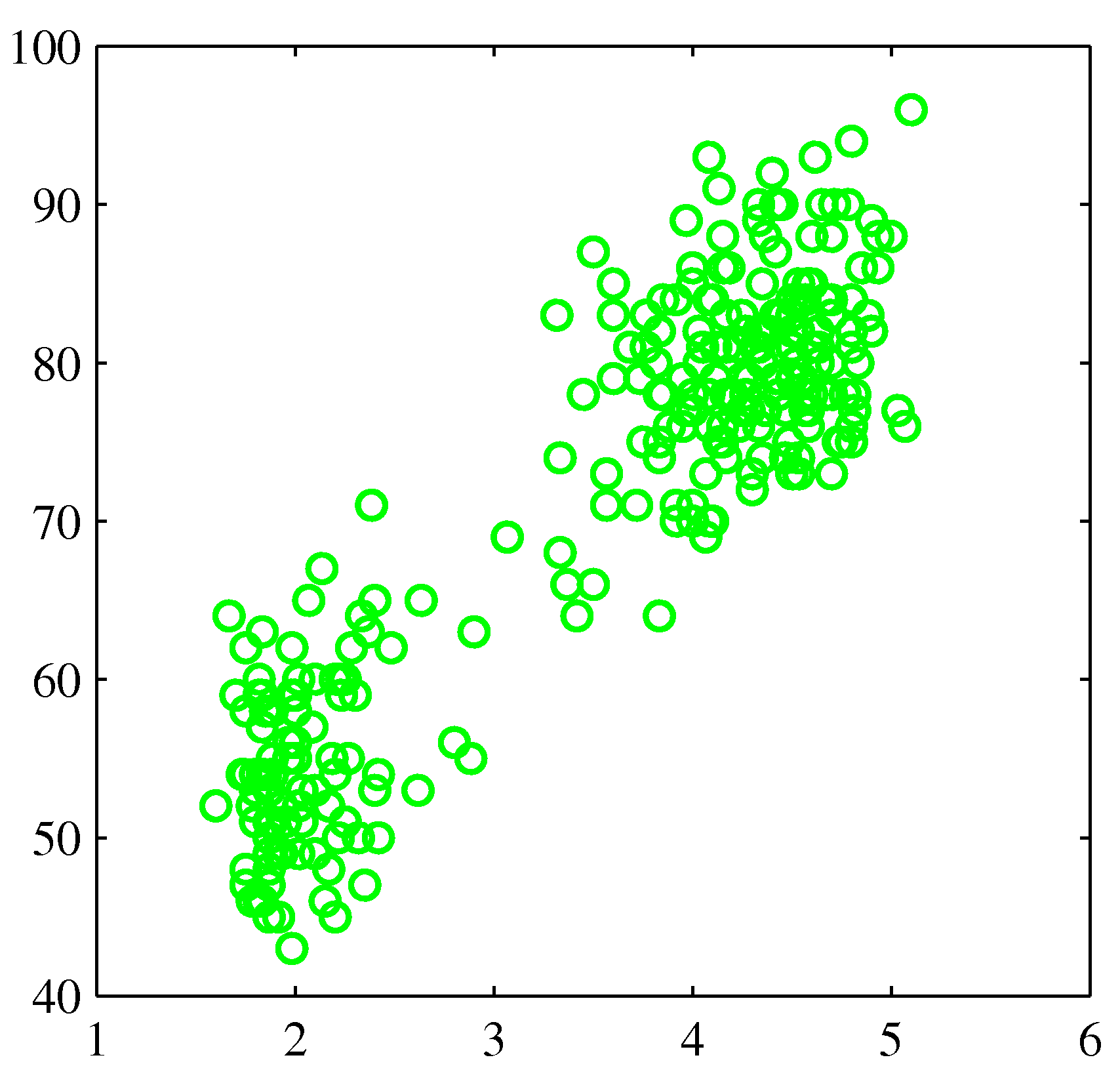
Illustrative Example¶
- You're now asked to build a density model for a data set (Old Faithful, Bishop pg. 681) that clearly is not distributed as a single Gaussian:
Unobserved Classes¶
Consider again a set of observed data $D=\{x_1,\dotsc,x_N\}$
- This time we suspect that there are unobserved class labels that would help explain (or predict) the data, e.g.,
- the observed data are the color of living things; the unobserved classes are animals and plants.
- observed are wheel sizes; unobserved categories are trucks and personal cars.
- observed is an audio signal; unobserved classes include speech, music, traffic noise, etc.
- Classification problems with unobserved classes are called Clustering problems. The learning algorithm needs to discover the classes from the observed data.
The Gaussian Mixture Model (GMM)¶
- If the categories were observed as well, these data could be nicely modeled by the previously discussed generative classification framework.
- Let us use equivalent model assumptions to linear generative classification:
where, as previously, we use notational shorthand $\mathcal{C}_k \triangleq (z_{nk}=1)$ and a hidden $1$-of-$K$ selector variable $z_{nk}$ with each observation $x_n$ as $$ z_{nk} = \begin{cases} 1 & \text{if } \, z_n \in \mathcal{C}_k\\ 0 & \text{otherwise} \end{cases} $$
- Since the class labels are not observed, the probability for observations in a GMM is obtained through marginalization over the classes (this is different in classification problems, where the classes are observed):
- The class priors $p(\mathcal{C}_k) =\pi_k$ are often called mixture coefficients.
Gaussian Mixture Models¶
- GMMs are very popular models. They have decent computational properties and are universal approximators of densities (as long as there are enough Gaussians of course)
Inference: Log-Likelihood for GMM¶
- The log-likelihood for observed data $D=\{x_1,\dotsc,x_N\}$,
... and now the log-of-sum cannot be further simplified.
- Compare this to the log-likelihood for $D=\{(x_1,y_1),\dotsc,(x_N,y_N)\}$ in (generative) classification:
which led to easy Gaussian and multinomial parameter estimation.
- Fortunately GMMs can be trained by maximum likelihood using an efficient algorithm: Expectation-Maximization.
Introducing a Soft Class Indicator¶
Let's introduce a $1$-of-$K$ selector variable $z_{nk}$ to represent the unobserved classes $\mathcal{C}_k$ by $$ z_{nk} = \begin{cases} 1 & \text{if $z_n$ in class $\mathcal{C}_k$}\\ 0 & \text{otherwise} \end{cases} $$
- We don't observe the class labels $z_{nk}$, but we can compute the posterior probability for the class labels, given the observation $x_n$:
- The $\gamma_{nk} \triangleq p(\mathcal{C}_k | x_n ) = p(z_{nk}=1 | x_n )$ are also called responsibilities. Note that $0 \leq \gamma_{nk} \leq 1$ by definition and that they can be evaluated (i.e., we can compute it).
- The responsibilities $\gamma_{nk}$ are soft class indicators and they play the same role in clustering as class selection variables ($y_{nk}$) do in classification problems.
ML estimation for Clustering: The Expectation-Maximization (EM) Algorithm Idea¶
- IDEA: Let's apply the (generative) classification formulas and substitute the reponsibilities $\gamma_{nk}$ wherever the formulas use the binary class indicators $y_{nk}$.
- Try parameter updates (same as for generative Gaussian classification):
- But wait ..., the responsibilities $\gamma_{nk}=\frac{\pi_k \mathcal{N}(x_n|\mu_k,\Sigma_k)}{\sum_j \pi_j \mathcal{N}(x_n|\mu_j,\Sigma_j)}$ are a function of the model parameters $\{\pi,\mu,\Sigma\}$ and the parameter updates depend on the responsibilities ...
- Solution: Iterate updating between $\gamma_{nk}$ and the parameters $\{\pi,\mu,\Sigma\}$. This procedure is called the Expectation-Maximization (EM) algorithm.
CODE EXAMPLE¶
We'll perform clustering on the data set from the illustrative example by fitting a GMM consisting of two Gaussians using the EM algorithm.
using DataFrames, CSV, LinearAlgebra
include("scripts/gmm_plot.jl") # Holds plotting function
old_faithful = CSV.read("datasets/old_faithful.csv")
X = convert(Matrix{Float64}, [old_faithful[1] old_faithful[2]]')
N = size(X, 2)
# Initialize the GMM. We assume 2 clusters.
clusters = [MvNormal([4.;60.], [.5 0;0 10^2]);
MvNormal([2.;80.], [.5 0;0 10^2])]
π_hat = [0.5; 0.5] # Mixing weights
γ = fill!(Matrix{Float64}(undef,2,N), NaN) # Responsibilities (row per cluster)
# Define functions for updating the parameters and responsibilities
function updateResponsibilities!(X, clusters, π_hat, γ)
# Expectation step: update γ
norm = [pdf(clusters[1], X) pdf(clusters[2], X)] * π_hat
γ[1,:] = (π_hat[1] * pdf(clusters[1],X) ./ norm)'
γ[2,:] = 1 .- γ[1,:]
end
function updateParameters!(X, clusters, π_hat, γ)
# Maximization step: update π_hat and clusters using ML estimation
m = sum(γ, dims=2)
π_hat = m / N
μ_hat = (X * γ') ./ m'
for k=1:2
Z = (X .- μ_hat[:,k])
Σ_k = Symmetric(((Z .* (γ[k,:])') * Z') / m[k])
clusters[k] = MvNormal(μ_hat[:,k], convert(Matrix, Σ_k))
end
end
# Execute the algorithm: iteratively update parameters and responsibilities
subplot(2,3,1); plotGMM(X, clusters, γ); title("Initial situation")
updateResponsibilities!(X, clusters, π_hat, γ)
subplot(2,3,2); plotGMM(X, clusters, γ); title("After first E-step")
updateParameters!(X, clusters, π_hat, γ)
subplot(2,3,3); plotGMM(X, clusters, γ); title("After first M-step")
iter_counter = 1
for i=1:3
for j=1:i+1
updateResponsibilities!(X, clusters, π_hat, γ)
updateParameters!(X, clusters, π_hat, γ)
iter_counter += 1
end
subplot(2,3,3+i);
plotGMM(X, clusters, γ);
title("After $(iter_counter) iterations")
end
PyPlot.tight_layout()
Note that you can step through the interactive demo yourself by running this script in julia. You can run a script in julia by
julia> include("path/to/script-name.jl")
Clustering vs. (Generative) Classification¶
| Classification | Clustering | |
| 1 | Class label $y_n$ is observed | Class label $z_n$ is latent |
| 2 | log-likelihood conditions on observed class $$\propto \sum_{nk} y_{nk} \log \mathcal{N}(x_n|\mu_k,\Sigma_k)$$ | log-likelihood marginalizes over latent classes $$\propto \sum_{n}\log \sum_k \pi_k \mathcal{N}(x_n|\mu_k,\Sigma_k)$$ |
| 3 | 'Hard' class selector $$ y_{nk} = \begin{cases} 1 & \text{if $y_n$ in class $\mathcal{C}_k$}\\ 0 & \text{otherwise} \end{cases} $$ | 'Soft' class responsibility $$\gamma_{nk} = p(\mathcal{C}_k|x_n)$$ |
| 4 | Estimation: $$\begin{align*} \hat{\pi}_k &= \frac{1}{N}\sum_n y_{nk} \\ \hat{\mu}_k &= \frac{\sum_n y_{nk} x_n}{\sum_n y_{nk}} \\ \hat{\Sigma}_k &= \frac{\sum_n y_{nk} (x_n-\hat\mu_k)(x_n-\hat\mu_k)^T}{\sum_n y_{nk}} \end{align*}$$ |
Estimation (1 update of M-step!) $$\begin{align*} \hat{\pi}_k &= \frac{1}{N}\sum_n \gamma_{nk} \\ \hat{\mu}_k &= \frac{\sum_n \gamma_{nk} x_n}{\sum_n \gamma_{nk}} \\ \hat{\Sigma}_k &= \frac{\sum_n \gamma_{nk} (x_n-\hat\mu_k)(x_n-\hat\mu_k)^T}{\sum_n \gamma_{nk}} \end{align*}$$ |
open("../../styles/aipstyle.html") do f
display("text/html", read(f,String))
end