Machine Learning¶
A Summary of lecture "Introduction to Computational Thinking and Data Science", via MITx-6.00.2x (edX)
- toc: true
- badges: true
- comments: true
- author: Chanseok Kang
- categories: [Python, edX, Machine_Learning]
- image: images/ml_block.png
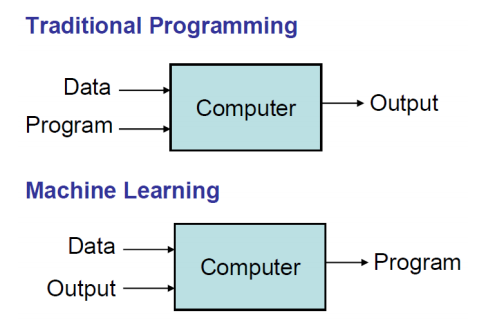
- What is Machine Learning
- Many useful programs learn something
Note: "Field of study that gives computers the ability to learn without being explicitly programmed" - Arthur Samuel
- Modern statistics meets optimization
- Basic Paradigm
- Observe set of examples: training data
- Infer something about process that generated that data
- Use inference to make predictions about previously unseen data: test data
- All ML Methods Require
- Representation of the features
- Distance metric for feature vectors
- Objective function and constraints
- Optimization method for learning the model
- Evaluation method
- Supervised Learning
- Start with set of feature vector / value pairs
- Goal : find a model that predicts a value for a previously unseen feature vector
- Regression models predict a real
- E.g. linear regression
- Classification models predict a label (chosen from a finite set of labels)
- Unsupervied Learning
- Start with a set of feature vectors
- Goal : uncover some latent structure in the set of feature vectors
- Clustering the most common technique
- Define some metric that captures how similar one feature vector is to another
- Group examples based on this metric
- Choosing Features
- Features never fully describe the situation
- Feature Engineering
- Represent examples by feature vectors that will facilitate generalization
- Suppose I want to use 100 examples from past to predict which students will pass the final exam
- Some features surely helpful, e.g., their grade on the midterm, did they do the problem sets, etc.
- Others might cause me to overfit, e.g., birth month
- Whant to maximize ratio of useful input to irrelevant input
- Signal-to-Noise Ratio (SNR)
- K-Nearest Neighbors
- Distance between vectors
- Minkowski metric
- Distance between vectors
In [1]:
from lecture12_segment2 import *
In [4]:
cobra = Animal('cobra', [1,1,1,1,0])
rattlesnake = Animal('rattlesnake', [1,1,1,1,0])
boa = Animal('boa\nconstrictor', [0,1,0,1,0])
chicken = Animal('chicken', [1,1,0,1,2])
alligator = Animal('alligator', [1,1,0,1,4])
dartFrog = Animal('dart frog', [1,0,1,0,4])
zebra = Animal('zebra', [0,0,0,0,4])
python = Animal('python', [1,1,0,1,0])
guppy = Animal('guppy', [0,1,0,0,0])
animals = [cobra, rattlesnake, boa, chicken, guppy,
dartFrog, zebra, python, alligator]
In [5]:
compareAnimals(animals, 3) # k=3
Help on method scale in module matplotlib.table:
scale(xscale, yscale) method of matplotlib.table.Table instance
Scale column widths by *xscale* and row heights by *yscale*.
- Using Distance Matrix for classification
- Simplest approach is probably nearest neighbor
- Remember training data
- When predicting the label of a new example
- Find the nearest example in the training data
- Predict the label associated with that example
- Advantage and Disadvantage of KNN
- Advantages
- Learning fase, no explicit training
- No theory required
- Easy to explain method and results
- Disadvantages
- Memory intensive and predictions can take a long time
- Are better algorithms than brute force
- No model to shed light on process that generated data
- Memory intensive and predictions can take a long time
- Advantages
In [6]:
# Applying scaling
cobra = Animal('cobra', [1,1,1,1,0])
rattlesnake = Animal('rattlesnake', [1,1,1,1,0])
boa = Animal('boa\nconstrictor', [0,1,0,1,0])
chicken = Animal('chicken', [1,1,0,1,2])
alligator = Animal('alligator', [1,1,0,1,1])
dartFrog = Animal('dart frog', [1,0,1,0,1])
zebra = Animal('zebra', [0,0,0,0,1])
python = Animal('python', [1,1,0,1,0])
guppy = Animal('guppy', [0,1,0,0,0])
animals = [cobra, rattlesnake, boa, chicken, guppy,
dartFrog, zebra, python, alligator]
In [7]:
compareAnimals(animals, 3) k = 3
Help on method scale in module matplotlib.table:
scale(xscale, yscale) method of matplotlib.table.Table instance
Scale column widths by *xscale* and row heights by *yscale*.
- A more General Approach: Scaling
- Z-scaling
- Each feature has a mean of 0 & a standard deviation of 1
- Interpolation
- Map minimum value to 0, maximum value to 1, and linearly interpolate
- Z-scaling
def zScaleFeatures(vals):
"""Assumes vals is a sequence of floats"""
result = np.array(vals)
mean = np.mean(vals)
result = result - mean
return result/np.std(result)
def iScaleFeatures(vals):
"""Assumes vals is a sequence of floats"""
minVal, maxVal = min(vals), max(vals)
fit = np.polyfit([minVal, maxVal], [0, 1], 1)
return np.polyval(fit, vals)
- Clustering
- Partition examples into groups (clusters) such that examples in a group are more similar to each other than to examples in other groups
- Unlike classification, there is not typically a "right answer"
- Answer dictated by feature vector and distance metric, not by a group truth label
- Optimization Problem
- Why not divide variability by size of cluster?
- Big and bad worse than small and bad
- Is optimization problem finding a $C$ that minimizes $dissimilarity(C)$?
- No, otherwise could put each example in its own cluster
- Need constraints, e.g.
- Minimum distance between clusters
- Number of clusters
- K-means Clustering
- Constraint: exactly k non-empty clusters
- Use a greedy algorithm to find an approximation to minimizing objective function
- Algorithm
randomly chose k examples as initial centroids
while true:
create k clusters by assigning each example to closest centroid
compute k new centroids by averaging examples in each cluster
if centroids don`t change:
break
In [8]:
from lecture12_segment3 import *
In [10]:
centers = [(2, 3), (4, 6), (7, 4), (7,7)]
examples = []
random.seed(0)
for c in centers:
for i in range(5):
xVal = (c[0] + random.gauss(0, .5))
yVal = (c[1] + random.gauss(0, .5))
name = str(c) + '-' + str(i)
example = Example(name, pylab.array([xVal, yVal]))
examples.append(example)
xVals, yVals = [], []
for e in examples:
xVals.append(e.getFeatures()[0])
yVals.append(e.getFeatures()[1])
random.seed(2)
kmeans(examples, 4, True)
Iteration #1 Cluster color = 0 Cluster with centroid [1.66014278 3.18525178] contains: (2, 3)-1 Cluster color = 1 Cluster with centroid [1.8494407 2.7367613] contains: (2, 3)-0, (2, 3)-2, (2, 3)-3, (2, 3)-4 Cluster color = 2 Cluster with centroid [5.57612073 6.33385138] contains: (4, 6)-0, (4, 6)-1, (4, 6)-2, (4, 6)-3, (4, 6)-4, (7, 7)-0, (7, 7)-1, (7, 7)-2, (7, 7)-3, (7, 7)-4 Cluster color = 3 Cluster with centroid [7.11402489 3.98797723] contains: (7, 4)-0, (7, 4)-1, (7, 4)-2, (7, 4)-3, (7, 4)-4 Iteration #2 Cluster color = 0 Cluster with centroid [1.49914988 3.08204521] contains: (2, 3)-1, (2, 3)-2, (2, 3)-4 Cluster color = 1 Cluster with centroid [2.28022797 2.44308067] contains: (2, 3)-0, (2, 3)-3 Cluster color = 2 Cluster with centroid [5.57612073 6.33385138] contains: (4, 6)-0, (4, 6)-1, (4, 6)-2, (4, 6)-3, (4, 6)-4, (7, 7)-0, (7, 7)-1, (7, 7)-2, (7, 7)-3, (7, 7)-4 Cluster color = 3 Cluster with centroid [7.11402489 3.98797723] contains: (7, 4)-0, (7, 4)-1, (7, 4)-2, (7, 4)-3, (7, 4)-4 Iteration #3 Cluster color = 0 Cluster with centroid [1.49914988 3.08204521] contains: (2, 3)-1, (2, 3)-2, (2, 3)-4 Cluster color = 1 Cluster with centroid [2.28022797 2.44308067] contains: (2, 3)-0, (2, 3)-3 Cluster color = 2 Cluster with centroid [5.57612073 6.33385138] contains: (4, 6)-0, (4, 6)-1, (4, 6)-2, (4, 6)-3, (4, 6)-4, (7, 7)-0, (7, 7)-1, (7, 7)-2, (7, 7)-3, (7, 7)-4 Cluster color = 3 Cluster with centroid [7.11402489 3.98797723] contains: (7, 4)-0, (7, 4)-1, (7, 4)-2, (7, 4)-3, (7, 4)-4
Out[10]:
[<lecture12_segment3.Cluster at 0x227612ac8c8>, <lecture12_segment3.Cluster at 0x227612ac408>, <lecture12_segment3.Cluster at 0x227612ac588>, <lecture12_segment3.Cluster at 0x227612ac6c8>]
- Mitigating Dependence on Initial Centroids
best = kMeans(points)
for t in range(numTrials):
C = kMeans(points)
if dissimilarity(C) < dissimilarity(best):
best = C
return best
- A Pretty Example
- User k-means to cluster groups of pixels in an image by their color
- Get the color associated with the centroid of each cluster, i.e., the average color of the cluster
- For each pixel in the original image, find the centroid that is its nearest neighbor
- Replaced the pixel by that centroid